Invited Symposium: SERCA-Type of Calcium Pumps and Phospholamban
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Results
It has been clearly demonstrated that several mRNA isoforms are generated by alternative splicing at the 3’end of SERCA 1 and SERCA 2 genes [1 for review]. Furthermore, in human platelets a novel SERCA 3 isoform recognized by the PL/IM 430 antibody but different from SERCA3 RK8-13 [3] was also identified [13]. Thus, we hypothesized that the SERCA 3 gene could also give rise to multiple isoforms by splicing at its 3’ end. In a first approach, we searched for the presence of multiple RNA populations by RNase Protection Assay (RPA). When we used as a probe a fragment of cDNA covering the 3’end of the coding sequence and the beginning of the untranslated region, two mRNA populations were observed, compatible with the existence of a divergency near the end of the coding region and thus with the presence of the two mRNA sequences published by Tokuyama et al. [14].
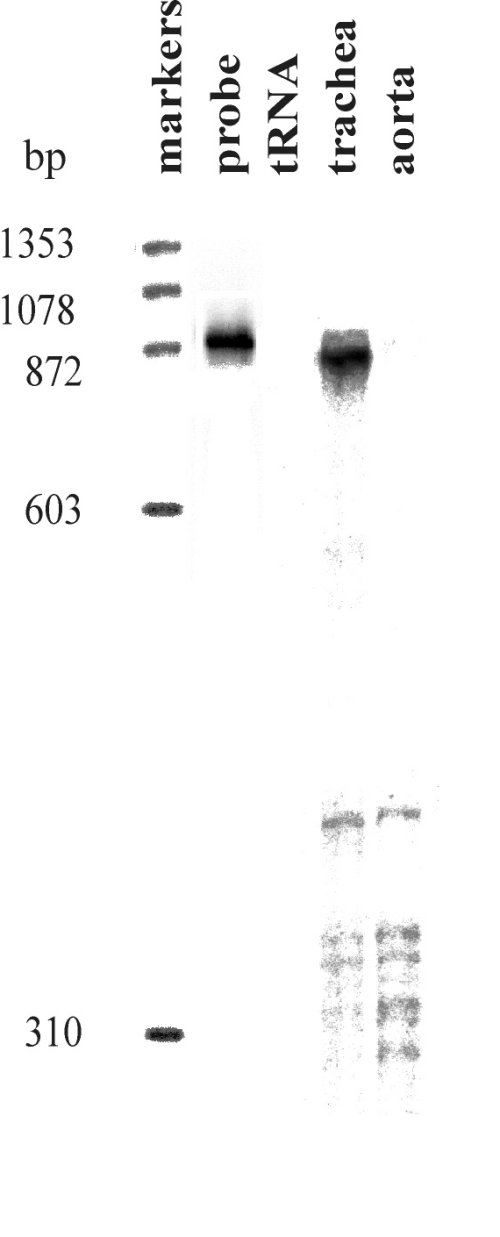 Fig. 1: RNase protection analysis of the expression of SERCA 3 mRNA variants.
Fig. 1: RNase protection analysis of the expression of SERCA 3 mRNA variants.
The fully protected (upper) fragment corresponding to SERCA 3a was present in trachea together with additional shorter fragments. Due to insertion of an additional exon in SERCA 3b, two major fragments of approximately 500bp and 300bp are expected for SERCA 3b. Only shorter fragments were present in RNA from aorta, corresponding to SERCA 3b.
To better characterize the mechanism of alternative splicing, we have isolated and sequenced the 3’end portion of the mouse SERCA 3 gene as shown in figure 2A
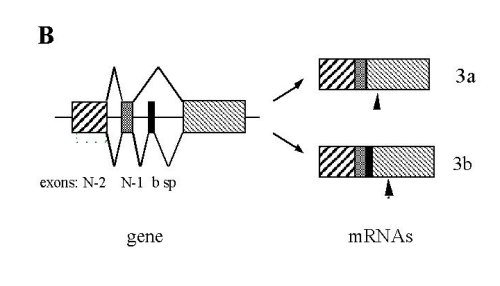
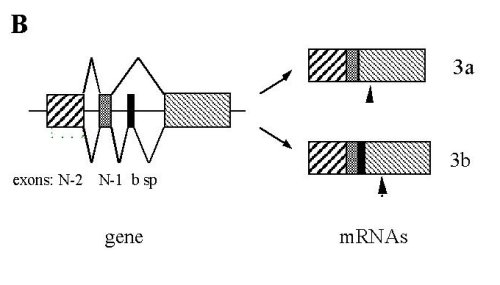
Fig. 2: Sequence and organization of the 3’end portion of the mouse SERCA 3 gene. .
A: Sequence of the fifth last exons (upper case) and introns (lower case). Nucleotides are numbered as referred [14]. Amino-acid sequence is presented beneath the nucleotide sequence with the one-letter code. Primers used for PCR are underlined: the upstream is 21S spanning from exons N-2 to N-1, the middle 21bS , and the downstream 22R. Bold letters and stars indicate the two different stop codons. B: Schematic representation of the 3’end of the mouse SERCA 3 gene and of the two mRNA species. Arrowheads indicate positions of stop codons.
We confirmed the sequence of SERCA 3b cDNA by sequencing a PCR product obtained by reverse transcription of cardiac RNA (not shown). The additional 73 nucleotides of SERCA 3b were located 387 nt downstream of the 3’ boundary of exon N-1 and constitute the b-specific exon (b sp). Exons were numbered by comparison with the human SERCA 3 gene [6] and each of them had consensus exon/intron boundaries. Addition of this exon modified the open reading frame of exon N and led to usage of two different stop codons. This splicing mechanism leads to the generation of two proteins which differ in their C-terminal portion by the substitution of the last 6 amino-acids of SERCA 3a by and extension of 45 amino-acids in SERCA 3b. We then studied distribution of the alternative transcripts in mouse tissues by RT-PCR using two sets of primers (see figure 2A for primers location).
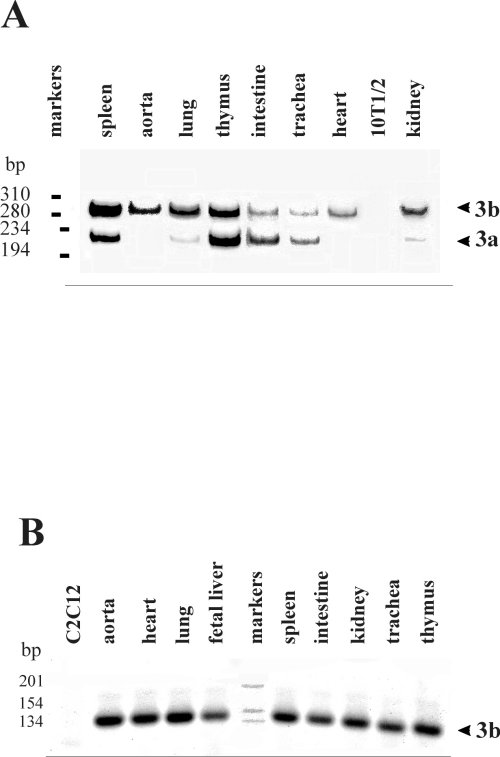 Fig. 3: RT-PCR analysis of SERCA 3 spliced variants expression. A: Southern-blot hybridization using an internal oligonucleotide of RT-PCR products obtained with primers 21S and 22R.. B: RT-PCR products stained with vistra green. Primers used were 21bS and 22R. Figure 3 is representative of at least 3 experiments performed on different RNA preparations from different mice. Positions of DNA markers are indicated.
Fig. 3: RT-PCR analysis of SERCA 3 spliced variants expression. A: Southern-blot hybridization using an internal oligonucleotide of RT-PCR products obtained with primers 21S and 22R.. B: RT-PCR products stained with vistra green. Primers used were 21bS and 22R. Figure 3 is representative of at least 3 experiments performed on different RNA preparations from different mice. Positions of DNA markers are indicated.
SERCA 3b was the only isoform expressed in aorta and heart and the major one in lung and kidney whereas the two isoforms were detected in all other tissues. No signal was detected in C2C12 skeletal muscle or 10T1/2 fibroblast cell lines and in controls without reverse transcriptase (not shown). The presence of SERCA 3b in aorta was in agreement with the presence of only short fragments in figure 1. Similarly, in trachea, SERCA 3a was slightly more abundant than SERCA 3b as shown by RPA and RT-PCR experiments (Figures. 1 and 3). We then used another pair of primers, 21bS and 22R, to detect specifically SERCA 3b or any other potential exon present between exon b sp and exon N. These results confirmed that SERCA 3b was expressed in each tissue and only one fragment was observed with these primers indicating that no other coding sequence was detectable between exons b sp and N.
Each RASMC expresses two SERCA isoforms and two Ca2+ release channels.
We used immunofluorescence and interferential contraste images to show that SERCA 2a and SERCA 2b were both expressed in each individual smooth muscle cell.
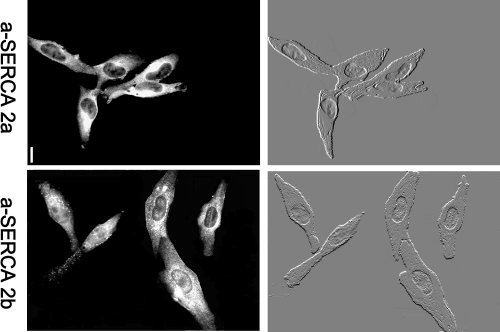 Fig.4 : Two calcium pumps are present in each differentiated RASMC in culture for 1 day. Comparison of immunolabeling and interferential contrast micrographs showed that each RASMC was labeled with each antibody. bars = 10 µm.
Fig.4 : Two calcium pumps are present in each differentiated RASMC in culture for 1 day. Comparison of immunolabeling and interferential contrast micrographs showed that each RASMC was labeled with each antibody. bars = 10 µm.
Proliferation is associated with loss of SERCA 2a and persistence of SERCA 2b.
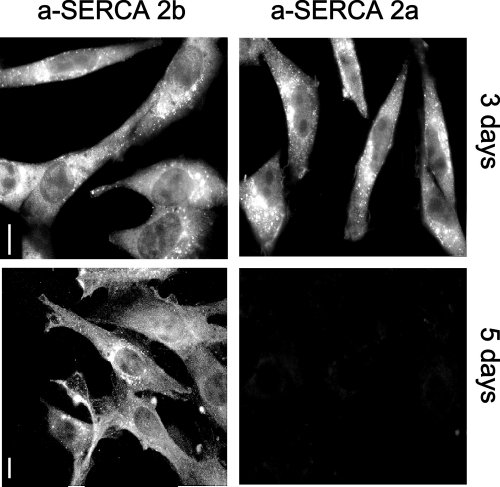 Fig.5 : Immunodetection of SERCA 2a and SERCA 2b in RASMC in culture for 3 or 5 days. On day 5, when cells were actively proliferating, SERCA 2a were not detected. Bar=10 µm.
Fig.5 : Immunodetection of SERCA 2a and SERCA 2b in RASMC in culture for 3 or 5 days. On day 5, when cells were actively proliferating, SERCA 2a were not detected. Bar=10 µm.
SERCA 2a and SERCA 2b were detected in RASMC after 3 days in culture. None of the cells were labeled with a-SERCA 2a on day 5 whereas all cells were labeled with a-SERCA 2b antibodies. The level of mRNA encoding the Ca2+ pumps was also studied by RT-PCR.
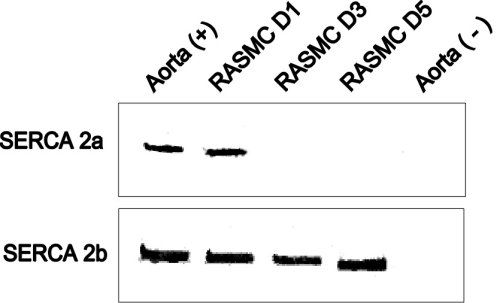 Fig.6 : Detection of SERCA 2 mRNA isoforms by RT-PCR in total aorta and RASMC in culture.
Fig.6 : Detection of SERCA 2 mRNA isoforms by RT-PCR in total aorta and RASMC in culture.
SERCA 2a and SERCA 2b mRNA isotypes were amplified from total RNA preparations from aorta, RASMC cultured for 1, 3, 5 days, and from control RNAs. SERCA 2a was present in RASMC cultured for one day but was absent on day 3 and 5.
Proliferation of RASMC is associated with loss of the caffeine-sensitive Ca2+ pool.
We performed experiments in the absence of external Ca2+ and in the presence of EGTA to eliminate any possible effect of ATP on Ca2+ influx and thus on CICR. At day 1, caffeine induced transient increases in [Ca2+]i. The stores were refilled by incubation in normal Ca2+ medium and an increase in [Ca2+]i was obtained when ATP was added.
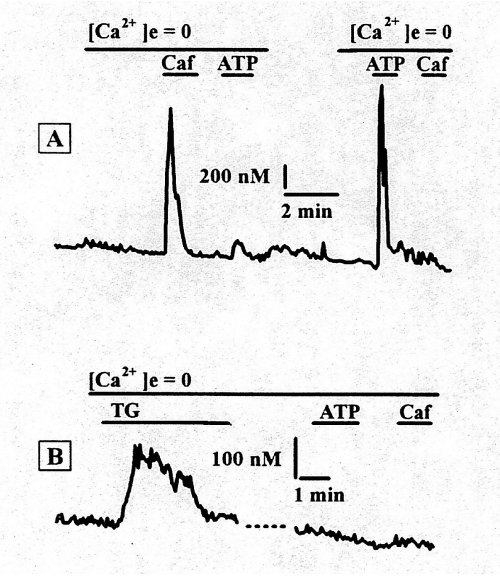 Fig.7A : Intracellular calcium transients in RASMCs in culture for 24h. Sequential application of 10 mM caffeine and 20 µM ATP. Fig.7B : The stores were refilled by incubation in normal calcium medium for 5 min, then 2 µM thapsigargin was added and incubated for 5 min in calcium free medium. Subsequent application of 20 µM ATP and 10 mM caffeine had no effect on intracellular calcium concentration.
Fig.7A : Intracellular calcium transients in RASMCs in culture for 24h. Sequential application of 10 mM caffeine and 20 µM ATP. Fig.7B : The stores were refilled by incubation in normal calcium medium for 5 min, then 2 µM thapsigargin was added and incubated for 5 min in calcium free medium. Subsequent application of 20 µM ATP and 10 mM caffeine had no effect on intracellular calcium concentration.
Incubation of the same cell with thapsigargin (Tg, 2 µM), a specific inhibitor of the SERCAs, for 10 min in Ca2+-free medium, resulted in a slow increase in the intracellular Ca2+ level causing depletion of the Ca2+ pools Neither caffeine nor ATP induced an increase in [Ca2+]i in the presence of Tg. Thus, the caffeine-sensitive and IP3-sensitive pools are both Tg-sensitive and contain a sarco(endo)plasmic reticulum Ca2+ pump.
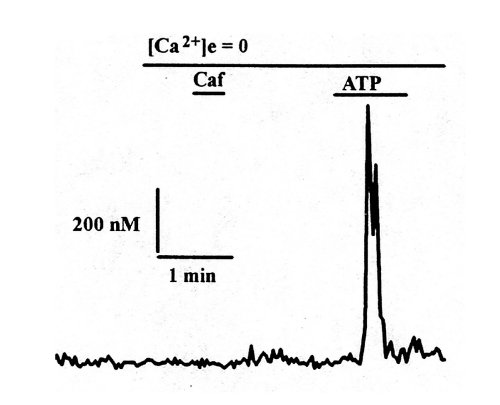 Fig.8 : Representative traces of calcium transients observed after application of caffeine (40 mM) and ATP (20 µM) in calcium-free medium, on Fura2-loaded RASMC in culture for 5 days.
Fig.8 : Representative traces of calcium transients observed after application of caffeine (40 mM) and ATP (20 µM) in calcium-free medium, on Fura2-loaded RASMC in culture for 5 days.
In figure 8 are presented representative traces of transient increases in [Ca2+]i observed after application of caffeine and ATP in Ca22+ free medium, in RASMC in culture for 5 days. On day 5, when RASMC were actively proliferating, caffeine did not induce Ca2+ release whereas a transient increase in [Ca2+]i was observed after application of ATP. This indicated that the cells had lost their functional caffeine-sensitive pool.
| <= Materials & Methods | RESULTS | Discussion & Conclussions => |
| Discussion Board | Next Page | Your Symposium |