Oxidative Stress Poster Session
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Results
·All clones presented in this poster have poly adenylation tails, a polyadenylation signal and a stop codon at the 3' end of the gene.
·None appear to be complete genes as reflected in the discrepancy between clone size and transcript size (see table 1).
·Open reading frames are absent so no translations are shown for the sequence data I have obtained so far.
Table 1: Summary of anoxic and frozen foot muscle clones obtained via differential screening of cDNA libraries.
LLAF = L. littorea Anoxic Foot
LLFF = L. littorea Frozen Foot
Clone Insert Transcript
Size Size (bases)
_________________________________
LLAF
RR 2050 1550
EE 1185 1350
DD 850 1350
7.1.4 645 1250
11.3 1066 1400
12.3.1 1000 1400
W 792 1000
10.1.1 950 1625
N 572 1100
__________________________________
LLFF
3 1106 ~1200
15 1651 ~6Kb
__________________________________
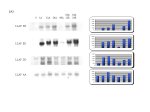
Figure 1: Up-regulated clones from the anoxia library that have similar, but not identical sequences. The sequence for these clones returned no hits through NCBI BLAST. However, FASTA returned a match for LLAF EE with glutenin, a protein found only in plants. The match has a score of 466 and a sum probability of 2.1e-44Shown are scans of the northern blots and histograms showing the relative band density.
(A) These results were obtained from the same set of northerns.
· Clone AA is included as an example of stable expression during the anoxia treatment.
· Clone EE has sequence homology with clone 7.1.4 and 11.3.
· Clone RR has not been sequenced, but is likely identical to clone EE because both were picked from the same tertiary screening plate and have the same expression pattern.
·Clone DD shows a similar expression pattern, but shows 2 bands, not one.
 click to enlarge
Figure 1A.
click to enlarge
Figure 1A.
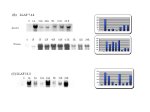
(B)The expression of clone LLAF 7.1.4 in response anoxia and freezing. Though the gene was isolated from the anoxic screen, it shows increased expression in response to freezing as well. Transcript levels peak at 1h of anoxia but not until 5 hours of freezing
.(C) Clone LLAF 11.3; the transcript level is very low in untreated animals (time 0) and shows variable expression during the stress.
 click to enlarge
Figure 1B and 1C
click to enlarge
Figure 1B and 1C
Figure 2: Sequence alignment of clones EE, 7.1.4, and 11.3.
These sequences are 60% identical and the alignment was obtained using DNAman. Sequences were obtained using automated procedures in both the forward and reverse directions to completion.
 click to enlarge
Figure 2
click to enlarge
Figure 2
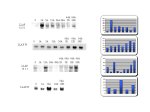
Figure 3: LLAF Clones that show increased expression in response to anoxia but no homology to other genes. On the left are the northern blot scans and on the right, Imagequant results of band density analysis
.
 click to enlarge
Figure 3
click to enlarge
Figure 3
The pattern of expression varies among these genes.
· LLAF 12.3.1 show strong expression after 1 h anoxia
· LLAF W shows gradual increase in expression throughout the time course.
· LLAF 10.1.1 shows variable expression over the time course, with the greatest level at 12 h anoxia treatment and during recovery.
· LLAF N also shows strongest expression at 12 hours anoxia.
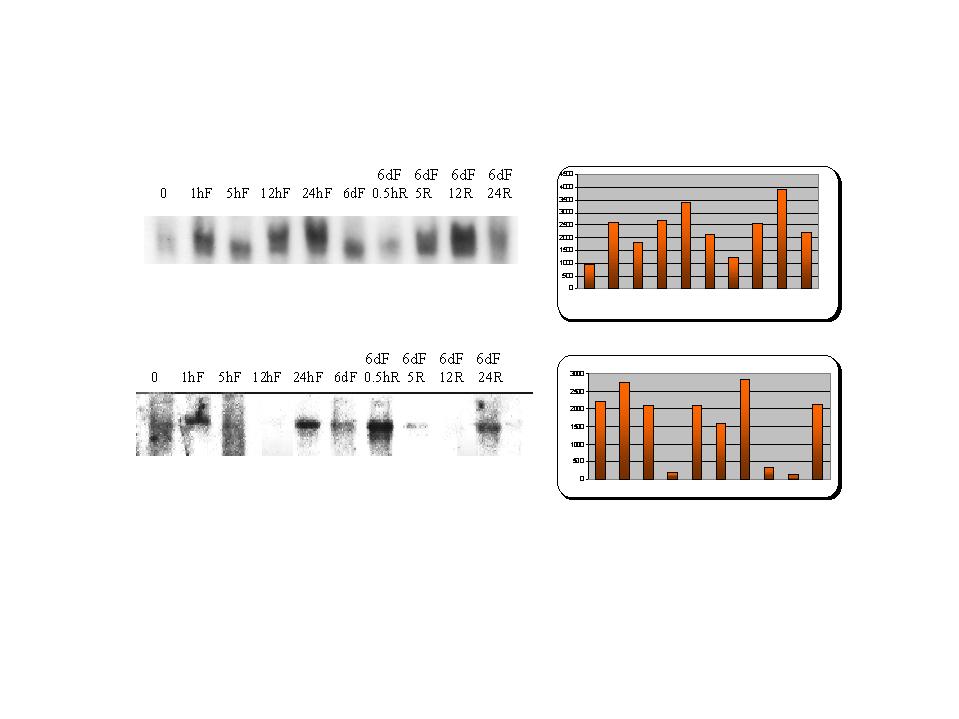
Figure 4: Northern blot analysis of upregulated clones obtained from the library screen of freeze-treated animals.
Both these genes show variable gene expression over the freezing time course. In spite of the less than stellear quality of the northerns it is clear that a trend of increasing transcript abundance is present.
 click to enlarge
Figure 4
click to enlarge
Figure 4
·Clone LLFF 3 has been sequenced but shows no homology with sequences in the BLAST database..
·Clone LLFF 15 has been sequenced from the forward and reverse directions to overlapping. The sequence is 1651 bases long and is heavy with stop codons from 365 bases on to the 3' end. Reading frame 1 is open to base 365 and BLAST hits indicate homology with myosin heavy chain. The BLAST alignment for this is shown in Figure 5. In addition, BLAST results show alignment of the beginning of clone 15 with the end of myosin. This alignment (if correct expected value of 7.8e-35) indicates that LLFF is truncated. This is further supported by the fact that the transcript size on the northern blot is approximately 6 Kb.
Figure 5. NCBI BLAST results for Clone LLFF 15; blastn, nr database.
gb|U59294|PMU59294 Placopecten magellanicus myosin heavy chain mRNA,complete cds. Length = 5826
Score = 356 (99.4 bits), Expect = 7.8e-35,
Identities = 105/150 (70%), Positives = 105/150 (70%), Strand = Plus / Plus
Query: 125 GRACAGGAACAGTNCATGCAGGTGGAGAAACAGCGCAAGGGCCTGGAACAGAAGGACAAG 184 | |||||| || || ||||| ||||| |||||| ||| ||| ||| |||| Sbjct: 5314 GAACAGGATCACAGCAACCAGGTCGAGAAGGTACGCAAGAACCTTGAAAGCCAGGTCAAG 5373
Query: 185 GACATGCAGGACAGACTGGACGAGGGCGAGGCTCAGGCACTCAAGGGAGGCAAGAAGARC 244 || | ||| | | |||| |||| || ||| |||||||||||||||||||| Sbjct: 5374 GAATTCCAGATCCGTTTGGATGAGGCTGAAGCTTCTTCACTCAAGGGAGGCAAGAAGCTA 5433
Query: 245 ATCCAGAAACWCGAACAGAGAGTGCGCGAG 274 |||||||||| ||| ||||||| |||| Sbjct: 5434 ATCCAGAAACTGGAATCCAGAGTGCACGAG 5463
Score = 265 (74.0 bits), Expect = 7.8e-35, Sum P(2) = 7.8e-35 Identities = 65/80 (81%), Positives = 65/80 (81%), Strand = Plus / Plus
Query: 277 GAGATGGAGCTTGCCAGCGAGCAGCGCCGACACGGGGAGACCCAGAAGAACATGCGCAAG 336 ||| || |||| || |||||| ||||| |||| |||||||||||||||||||||||| Sbjct: 5467 GAGGCTGAACTTGACAACGAGCAACGCCGTCACGCCGAGACCCAGAAGAACATGCGCAAG 5526
Query: 337 GCCGACAGACGCGTCAAGGA 356 |||||| | || |||| || Sbjct: 5527 GCCGACCGTCGTCTCAAAGA 5546
| <= Materials & Methods | RESULTS | Discussion & Conclussions => |
| Discussion Board | Next Page | Your Poster Session |