Invited Symposium: Molecular Physiology of Sodium-Calcium Exchange
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Introduction
The Na
Quantitative end-labeled RT-PCR analysis (QERT-PCR).
A pair of oligonucleotides flanking the alternatively spliced region of NCX1 (nts 1823-1842) and (nts 2105-2124) (9) were used for QERT-PCR. One oligonucleotide was end-labeled and used in PCR with cDNA prepared from RNA obtained from cultured cells. Products were removed after different numbers of amplification cycles and fractionalted using a 5% polyacrylamide sequencing gel. Dried gels were analyzed using a phosphoimage system and ImageQuant (Molecular Dynamics) software. This technique is described in detail elsewhere (8).
We demonstrated by mixing different ratios of amplifiable material that the size of the amplified product (from 109 bp to 193 bp) did not affect the amount of PCR products (data not shown). We also observed that the amount of PCR product routinely differed by less than 3% from the expected to be found in the admixing experiments.
Functional studies in Xenopus oocytes.
The 2909 bp full-length rat cardiac NCX1 cDNA was used to construct different full-length NCX1 isoforms into pSD64TF vector (10). SP6 mMessage mMachine kit (Ambion) was used to synthesize the complementary RNA from clones containing either AD or BD isoforms of rat NCX1. Oocytes were harvested by surgery, isolated using collagenase and maintained at 16C as described (11). cRNA was injected into stage V and VI oocytes and after two days Na Chimeric constructs between exons A and B were made by PCR and gene sewing as described by Horton et al. (14). These novel fragments are cloned into the full-length NCX1 gene in the pSD64 vector so cRNA can be synthesized and used in Xenopus oocytes to study NCX1 function.
Quantitative Analysis of isoforms of NCX1
We used the QERT-PCR procedure to rapidly analyze the relative amount of the different isoforms present in RNA. Amplification was performed with one of the primers end-labeled. When the PCR products are separated on a sequencing gel, the size permits identification of the composition of the isoform and the band intensity reflects the relative amount of that isoform. By detecting the products in this manner, only small amount of cDNA and limited number of amplification cycles are required. Preliminary experiments by mixing various ratios of plasmid DNA from the different astrocyte clones had shown that the amounts of QERT-PCR products correlated with the relative amounts of DNA and there was no bias on amplification based on the size of the products (see Methods).
An example of the QERT-PCR technique is shown (Fig. 1). Rat astrocyte RNA was used to prepare cDNA by using either oligo dT or random primers. The cDNA is amplified using primers that flank the alternatively spliced region of NCX1. One of the primers was end-labeled with 32P prior to the reaction. When products are removed after different number of cycles and fractionated on the sequencing gel you can see a clear increase in specific product. For astrocytes three predominant bands are clearly visible. Those bands represent isoforms containing exons BDEF, BDF and BD all previously identified by cloning and sequencing of the PCR products from astrocyte cDNA (8).
Quantitation of NCX1 isoforms in astrocyte and neuronal cultures
This procedure was used to identify and quantitate the NCX1 isoforms present in astrocyte RNA. We observed that for two independent analyses of astrocyte RNAs three predominant bands of near equal intensity can be observed (Fig. 1A, lane 1 for 20 cycles and lane 2 for 23 cycles). The size of these bands correspond to BDEF, BDF and BD isoforms. When the bands for 23 cycles of amplification (lane 2) were quantitated using the phosphoimager method, the three predominant isoforms each represented about 23% of the total labeled product (Fig. 2A). Similar results were obtained when another group of neonatal rats was used to culture astrocytes and the amplification was repeated. These results demonstrate that three predominant isoforms of equal amounts constitute most of the NCX1 message in primary astrocytes. We show that the same three isoforms are expressed in the astrocyte-like cell line C6 (8).
To investigate whether different brain cell types utilize specific NCX1 isoforms, we performed QERT-PCR on rat primary hippocampal neurons. There were two predominant isoforms present in neurons and they differed from those in the astrocytes and the size suggested that the two isoforms were ADF and AD. Sequence analysis confirmed the identification of the bands as ADF and AD. Other minor bands accounted for less than 5% of total signal intensity in the lane of 23 cycles. Therefore, the major isoforms for the astrocytes and neurons were different with the astrocytes utilizing exon B-containing isoforms and the neurons, exon A-containing isoforms.
Activation of PKA and function of NCX1 isoforms
To determine if there is any functional difference between the A and B exon containing isoforms, we cloned the full-length rat Na Oocytes expressing the BD isoform displayed a Na
Further mapping of regions affecting PKA activation of the NCX
We show that A exon containing isoforms like AD increase their function after activation of the PKA pathway whereas the BD isoform does not. To further map the portion of the A exon that is responsible for this increase in function we have made chimeras between the A and B exons and have placed them in a full-length construct. RNA from these constructs have been used to study NCX1 function in Xenopus oocytes.
Preliminary studies have demonstrated that chimeric molecules do not affect NCX1 function. Interestingly, only those chimeras that contain the 3' half of exon A are increased in function after study in the oocyte expression system.
The Na The results presented demonstrate that the isoforms of NCX1 function differently. This supports our hypothesis that isoforms of NCX1 can be regulated in a cell-specific manner. For example, a number of neurotransmitters in brain have been shown to induce phosphorylation of membrane proteins. When the neurotransmitters are released in the vicinity of neurons, the neurotransmitter may selectively enhance the activity of the neuronal exchanger by phosphorylation.
1. Philipson KD, Nicoll, DA, Matsuoko, S, Hryshko, LV, Levitsky, DO, Weiss, JN (1996) Ann N Y Acad Sci 779:20-28.
2. Ruknudin A, Valdivia C, Kofuji P, Lederer WJ, Schulze DH (1997) Na/Ca exchanger in Drosophila: cloning, expression, and transport differences. Am J Physiol 273:C257-C265
3. He, Z, Tong, Q, Quednau, BD, Philipson, KD, Hilgemann, DW ( 1998) Cloning, expression and characterization of the Squid Na-Ca exchanger (NCX-SQ1). J Gen Physiol 111:857-873
4. Linck, B, Qui,Z, He, Z, Tong, Q, Hilgemann, DW, Philipson, KD. (1998) Functional comparison of the three isoforms of the Na/Ca exchanger (NCX1, NCX2 and NCX3). Am J Physiol 274:C415-C423.
5. Kofuji P, Lederer WJ, Schulze DH (1994) Mutually exclusive and cassette exons underlie alternatively spliced isoforms of the Na/Ca exchanger. J Biol Chem 269:5145-5149.
6. Kofuji P, Hadley RW, Kieval RS, Lederer WJ, Schulze DH (1992) Expression of the Na-Ca exchanger in diverse tissues: a study using the cloned human cardiac Na-Ca exchanger. Am J Physiol 263:C1241-C1249.
7. Lee SL, Yu ASL, Lytton J (1994) Tissue-specific expression of Na-Ca exchanger isoforms. J Biol Chem 269:14849-14852.
8. He,S. et al. (1998) Isoform-specific regulation of the Na/Ca exchanger in rat astrocytes and neurons by PKA. J. Neuroscience 18:4833-4841.
9. Low W, Kasir J, Rahamimoff H (1993) Cloning of the rat heart Na-Ca exchanger and its functional expression in HeLa cells. FEBS Lett 316:63-67.
10. Krieg PA, Melton DA (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nuc Acid Res 12:7057-7070.
11. Goldin AL (1992) Maintenance of Xenopus laevis and oocyte injection. Methods Enzymol 207:266-279.
12. Kuzhikandathil EV, Molloy GR (1994) Transcription of the brain creatine kinase gene in glial cells is modulated by cyclic AMP-dependent protein kinase. J Neurosci Res 39:70-82.
13. Kamei C, Mukai T, Tasaka K (1992) Histamine-induced depolarization and the cyclic AMP-protein kinase A system in isolated guinea pig adipocytes. Jpn J Pharmacol 60:179-186.
14. Horton, RM, Hunt, HD, Ho, SN, Pullen, JK, Pease, LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:69-77.
Materials and Methods
Results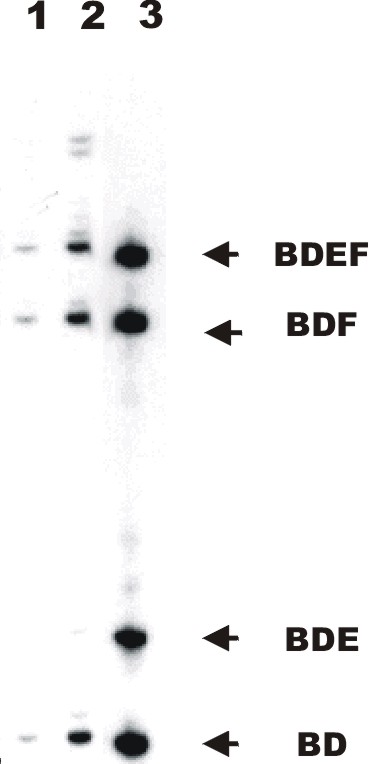
Fig1. QERT-PCR analysis on RNA from primary astrocytes. Astrocyte cDNA was amplified by a pair of oligonucleotides annealing to 5' and 3' common region. Same volume of QERT-PCR product at different cycle numbers (cycle 20 and 23) was loaded into 5% sequencing gel and separated (lanes 1 and 2 respectively). Lane 3 is the PCR product from a 1:1:1:1 mixture of plasmid DNA of four-representative astrocyte cloned NCX1 isoforms previously sequenced. The isoform designation is presented to the right of the figure.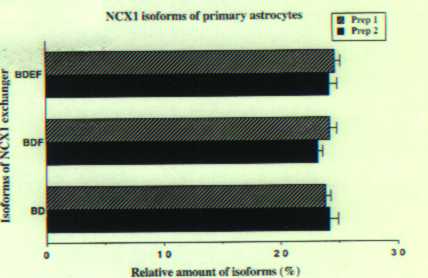 Fig2A. Analysis of QERT-PCR of major NCX1 isoforms in rat cultured cells. Signal intensity of each NCX1 isoform from was analyzed and relative amount of the predominant isoforms were plotted as the percentage of total signal intensity. Values are Means +/- S.E.M. from two experiments. A. cultured astrocytes (See Fig. 1). B. cultured hippocampal neurons...
Fig2A. Analysis of QERT-PCR of major NCX1 isoforms in rat cultured cells. Signal intensity of each NCX1 isoform from was analyzed and relative amount of the predominant isoforms were plotted as the percentage of total signal intensity. Values are Means +/- S.E.M. from two experiments. A. cultured astrocytes (See Fig. 1). B. cultured hippocampal neurons...
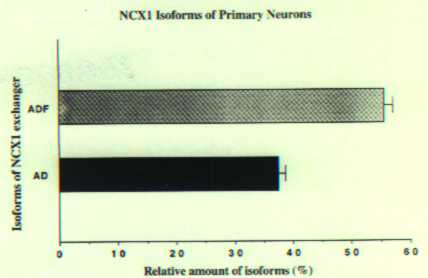 Fig2B.
Fig2B.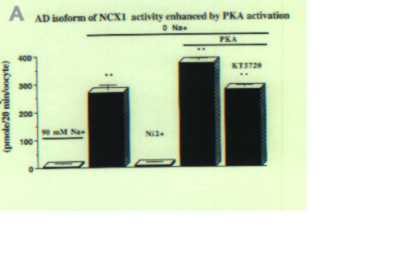
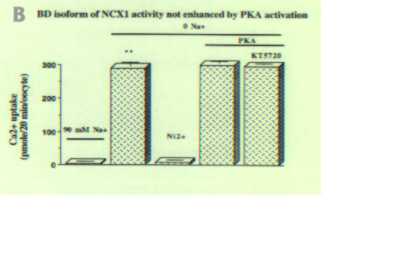
Fig3. PKA activation increases Ca2+ influx of neuron NCX1 isoform but not astrocyte isoform. A. 45Ca2+ influx was measured in Xenopus oocytes injected with cRNA of neuron isoform AD. Oocytes were exposed to 90 mM NaCl or 90 mM KCl (0 Na). 5 mM NiCl2 was used to block the exchanger. Ca2+ uptake in 0 Na + solution was significantly higher than in 90 mM Na + solution (p>0.001) and was blocked by Ni2+. For PKA activation experiment, a PKA cocktail was included in the 90 mM NaCl pre-incubating solution. 1 µM KT5720 was used to block PKA activation. After PKA activation, Ca2+ uptake was significantly higher than in 0 Na+ solution (p>0.001) and upregulation was blocked by KT5720 significantly (p>0.001). Values are Means ± S.E.M. and each Mean value contains results from 10 to 25 oocytes. B. 45 Ca2+ influx was measured in Xenopus oocytes injected with cRNA of astrocyte isoform BD as described in A
Discussion and Conclusion
References
| Discussion Board | Previous Page | Your Symposium |