Invited Symposium
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Introduction
SNARE proteins have recently been implicated in numerous vesicle traffic events. This superfamily of proteins was originally identified in neuronal tissues as components of the docking and fusion machinery of synaptic vesicles with the presynaptic membrane. Three neuronal SNARE proteins, syntaxin-1a, VAMP-2 and SNAP25, likely form a highly stable ternary complex as a major step in neurotransmitter exocytosis. This complex is stable in denaturing detergents such as sodium dodecyl sulfate and the core of the complex is resistant to digestion by endoproteases. In contrast to the focus on neuronal SNARE proteins, much less is known about the ability of other combinations of SNARE proteins to form ternary or higher order complexes.
It is well documented that membranes containing the GLUT4 glucose transporter of muscle and fat cells are translocated to the cell surface in response to insulin and it is thought that SNARE proteins play a central role in this translocation. Of the three most important neuro nal SNARE proteins, VAMP-2, SNAP25 and syntaxin- 1a, only VAMP-2 is found in cells which regulate their glucose uptake in response to insulin. Instead, these cells express other SNARE isoforms, specifically syntaxin-4 and SNAP23. Recent work from our lab and others has implicated syntaxin-4 and VAMP-2 in the process of insulin-stimulated GLUT4 translocation (1, 2, 3, 4).
Here we present data demonstrating that SNAP23 binds both syntaxin-4 and VAMP-2 specifically and with high affinity. However, these three proteins, in contrast to the subset of neuronal SNAREs, SNAP25, VAMP-2 and syntaxin-1a, do not form a stable, SDS-resistant complex.
We have also tested the possible participation of SNAP23 in GLUT4 traffic by introduction of antibodies or recombinant proteins into 3T3-L1 adipocytes which could interfere with or mimic SNAP23 function. The results show that microinjected antibodies to the C-terminal domain of SNAP23 reduce the insulin-dependent GLUT4 incorporation into the plasma membrane. T hese observations suggest that SNAP23 is required for vesicle-membrane interaction. We further show that full length SNAP23 increases insulin action on GLUT4 and glucose uptake in microinjected and chemically permeabilized cells, and propose that SNAP23 may be limiting for insulin-dependent GLUT4 incorporation into the membrane. This makes SNAP23 a likely target for regulation.
Materials and Methods
Fusion proteins and antibodies.
Polyclonal antibodies to GLUT4 (5), SNAP23 (termed anti-SN23.c12) (6), syntaxin-4 (termed anti-Sy4) (3), and VAMP-2 (termed anti-VAMP-2) (5) were prepared previously. Fluorescein isothiocyanate-conjugated donkey-anti-rabbit antibodies and rhodamine-conjugated dextran were obtained from Molecular Probes, Inc. (Eugene, OR). Recombinant fusion proteins were prepared as described previously (6, 7). Horseradish peroxidase-conjugated protein A secondary antibodies were purchased from Bio-Rad.
In vitro binding assays.
Binding assays were performed following the procedure of Pevsner et. al, (1994) for neuronal SNAREs. Briefly, recombinant GST-SNARE fusion proteins (termed the ‘fixed’ SNARE) was bound to glutathione agarose beads and mixed with soluble recombinant SNARE(s) cleaved from GST as described above. The proteins were incubated in 200 µl of binding buffer (5 mM HEPES pH 7.4, 70 mM KCl, 1 mM MgCl2, and 0.25% Triton-X-100) for 2 h at 4°C with e
nd-over-end rotation. The beads were collected by a low speed centrifugation (10 s, ~1000xg) and washed twice with 1 ml binding buffer. Twenty µl of 2x SDS sample buffer (125 mM Tris-HCl pH 6.8, 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue) were added and each tube was boiled for 2-3 minutes for separation by SDS-PAGE on 14% polyacrylamide gels.
SDS-resistant complexes.
Rat brain homogenate solublized in Triton-X-100 was prepared as described (8). 3T3-L1 mouse adipocyte crude membranes were prepared as described (6). The SDS-resistance of endogenous SNARE complexes was assayed using 20 µg of Triton X-100 solubilized rat brain protein or 90 µg of Triton X-100 solubilized 3T3-L1 mouse adipocyte crude membrane protein incubated at 4°C for 2 h. The SDS-resistance of recombinant protein complexes was tested using equimolar amounts of the recombinant forms of VAMP-2, SNAP25, SNAP23, syntaxin-1 and syntaxin-4, all cleaved of GST. The proteins were incubated for 2 h at 4
°C in Binding buffer. 2x SDS-sample buffer was added and some of the samples were boiled for 3 min.
Single cell microinjection and PM lawns.
3T3-L1 cells in a marked, 1 mm2 region of the coverslip were microinjected as described previously (9). Approximately 90% of the cells in the marked region (~100 cells) were microinjected with 20 µM GST or GST-SNAP23 or 0.1 µg/µL anti-SN23.c12 or irrelevant IgG in a solution containing 1.1 µg/µL rhodamine-dextran (Mr 10,000), 110 mM potassium acetate, 10 mM HEPES (pH 7.2) and 1 mM EDTA. The volume microinjected was about 1/10 of the cell volume. After microinjection cells were incubated in DMEM for 2 hours followed by a 15 min stimulation with 100 nM insulin, where indicated. Plasma membrane lawns (sheets) were prepared as described by Volchuk, et al. (9). Fluorescent images were obtained using a Leica inverted fluorescence microscope (model DM IRB) and quantitated and quantitated and analyzed as described by Volchuk, et al. (9).
Glucose transport in SLO-permeabilized cells.
The ability of insulin to stimulate glucose transport in SLO-permeabilized 3T3-L1 adipocytes was measured as described previously (3). Following permeabilization the SLO solution was removed and replaced with potassium glutamate buffer containing an ATP-regeneration system and 20 µM of either GST-SNAP23 or GST and incubated for 15 min at 37°C. Insulin (100 nM), where indicated, was added for a further 15 min in the presence of the proteins. Stock transport solution (final concentrations: 10 µM [3H]2-deoxyglucose (1 µCi/mL), 2 µM [14C]sucrose (0.2 µCi/mL)) was then added directly to the wells and incubated for a further 5 min at room temperature. Statistical analysis was done using ANOVA.
Results
Binding assays were performed to quantitate binary interactions between SNAP23 and its two putative endogenous binding partners VAMP-2 and syntaxin-4. Immobilized GST-VAMP-2 and GST-syntaxin-4 (0.3 µM) were titrated with increasing concentrations of SNAP23. Specific and saturable binding of SNAP23 to the fixed SNAREs was observed for both binary interactions (Fig 1). The syntaxin-4:SNAP23 interaction was the stronger of the two with an apparent half-maximal binding coefficient of 0.76 ± 0.15 µM (Fig. 1a). SNAP23 bound to VAMP-2 with an apparent half-maximal binding coefficient of 2.8 ± 0.1 µM (Fig. 1b). Since all of the recombinant proteins may not be in a fully native conformation, these values represent the minimum binding affinities of the individual SNARE proteins.
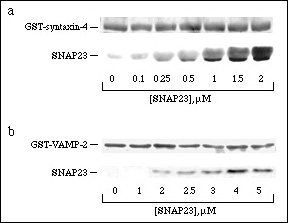 Figure 1. Binary binding of SNAP23, syntaxin-4 and VAMP-2. Increasing concentrations of SNAP23, were used to titrate 0.3 µM GST-syntaxin-4 (a) or 0.3 µM GST-VAMP-2 (b) to saturation. Glutathione-agarose was used to sediment SNAP23 bound to syntaxin-4 or VAMP-2. The pellets were analyzed by 14% SDS-PAGE and transferred to nitrocellulose. Ponceau S was used to visualize the GST constructs (i.e. GST-syntaxin-4 and GST-VAMP-2) while SNAP23 was immunoblotted with anti-SN23.C12.
Figure 1. Binary binding of SNAP23, syntaxin-4 and VAMP-2. Increasing concentrations of SNAP23, were used to titrate 0.3 µM GST-syntaxin-4 (a) or 0.3 µM GST-VAMP-2 (b) to saturation. Glutathione-agarose was used to sediment SNAP23 bound to syntaxin-4 or VAMP-2. The pellets were analyzed by 14% SDS-PAGE and transferred to nitrocellulose. Ponceau S was used to visualize the GST constructs (i.e. GST-syntaxin-4 and GST-VAMP-2) while SNAP23 was immunoblotted with anti-SN23.C12.
One novel property of the neuronal SNARE complex of SNAP25, syntaxin-1 and VAMP-2 is its ability to withstand denaturation by SDS (10). It is not known whether SNAP23, syntaxin-4 and VAMP-2 can form such a complex or whether SNAP23 or syntaxin-4 can substitute for their cognate neuronal SNARE in the complex. We therefore examined the ability of SNAP23 and syntaxin-4 to participate in SDS-resistant complexes.
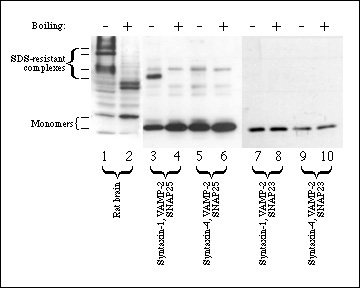 Figure 2. Only neuronal SNARE complexes are SDS-resistant. Triton-X-100 solubilized rat brain microsomes (lanes 1-2) were run as a positive control for SDS-resistant complexes. Equimolar concentrations (3.3 µM) of VAMP-2 (lanes 3-10), SNAP25 (lanes 3-6), SNAP23 (lanes 7-10), syntaxin-1a (lanes 3-4,7-8) and syntaxin-4 (lanes 5-6, 9-10) were incubated for 2 h at 4°C to allow complexes to form. SDS sample buffer similar in composition to SDS-PAGE buffer (see Methods) was added to all samples and samples were either boiled (lanes 2, 4, 6, 8, 10) or not boiled (lanes 1, 3, 5, 7, 9). Samples were analyzed by 12% SDS-PAGE, transferred to nitrocellulose and immunoblotted with syntaxin-1 antibody (lanes 1-2), SNAP25 antibody (lanes 3-6) or SNAP23 antibody (lanes 7-10) as described in Methods. 60 kDa bands, indicated in lanes 3-6 by an arrowhead, are non-specific immunoreactive contaminants.
Figure 2. Only neuronal SNARE complexes are SDS-resistant. Triton-X-100 solubilized rat brain microsomes (lanes 1-2) were run as a positive control for SDS-resistant complexes. Equimolar concentrations (3.3 µM) of VAMP-2 (lanes 3-10), SNAP25 (lanes 3-6), SNAP23 (lanes 7-10), syntaxin-1a (lanes 3-4,7-8) and syntaxin-4 (lanes 5-6, 9-10) were incubated for 2 h at 4°C to allow complexes to form. SDS sample buffer similar in composition to SDS-PAGE buffer (see Methods) was added to all samples and samples were either boiled (lanes 2, 4, 6, 8, 10) or not boiled (lanes 1, 3, 5, 7, 9). Samples were analyzed by 12% SDS-PAGE, transferred to nitrocellulose and immunoblotted with syntaxin-1 antibody (lanes 1-2), SNAP25 antibody (lanes 3-6) or SNAP23 antibody (lanes 7-10) as described in Methods. 60 kDa bands, indicated in lanes 3-6 by an arrowhead, are non-specific immunoreactive contaminants.
Figure 2 confirms previous observations that syntaxin-1 and SNAP25 form an SDS-resistant complex with VAMP-2. These complexes could be detected in homogenized brain tissue or in mixtures of recombinant proteins. In contrast however, substitution of either syntaxin-1 or SNAP25 with their respective non-neuronal counterparts did not allow formation of an SDS-resistant complex (Fig. 2). In addition, no detectable complex between SNAP23, syntaxin-4 and VAMP-2 is found (Fig. 2). No SDS-resistant complexes were seen in solubilized crude membrane fractions from 3T3-L1 adipocytes (results not shown), possibly due to the low abundance of SNARE proteins in these cells relative to neuronal cells.
To examine the participation of endogenous SNAP23 in insulin-dependent incorporation of GLUT4 into the cell membrane, antibodies raised to a hemocyanin-linked peptide comprising the C-terminal 12 amino acids of SNAP23 (termed anti-SN23.c12) were microinjected into 3T3-L1 adipocytes. Digital quantitation of lawns from four independent experiments demonstrated that microinjection of anti-SN23.c12 prevented the insulin-stimulated increase in GLUT4 on the membrane lawns (statistically significant, p<0.05) (Fig. 3). Unrelated IgG did not have any effect on insulin action.
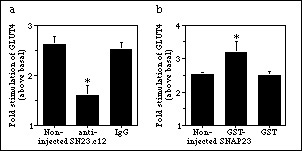 Figure 3. Effect of SNAP23 antibodies on insulin effects. Fluorescent images of GLUT4 in plasma membrane lawns were digitally quantitated by measuring average intensities for individual lawns for antibody (a) or fusion protein-injected (b) cells. Values from individual experiments were averaged and normalized to basal values. Values represent mean ± SE of the effect of insulin relative to its corresponding basal control, of five independent experiments. (*) denotes data which are significantly different from either control (p < 0.05, ANOVA).
Figure 3. Effect of SNAP23 antibodies on insulin effects. Fluorescent images of GLUT4 in plasma membrane lawns were digitally quantitated by measuring average intensities for individual lawns for antibody (a) or fusion protein-injected (b) cells. Values from individual experiments were averaged and normalized to basal values. Values represent mean ± SE of the effect of insulin relative to its corresponding basal control, of five independent experiments. (*) denotes data which are significantly different from either control (p < 0.05, ANOVA).
To more directly prove the participation of SNAP23 in GLUT4 organelle fusion, we tested the effect of exogenous full length SNAP23 in this process. The protein is naturally soluble but attaches to membranes via palmitoylation and association with other proteins such as syntaxin-4. Therefore, it was conceivable that introduction of an excess full-length SNAP23 might increase membrane fusion events if free syntaxin-4 and incoming vesicles are present.
GST-SNAP23 was microinjected into a clearly identified area of serum-deprived 3T3-L1 adipocytes. Based on the volume microinjected and the calculated cell volume, the final concentration of SNAP23 introduced may reach 2 µM. The GLUT4 signal in the non-injected cells was assigned a value of 1.00 within each experiment, and the effect of GST-SNAP23 was calculated relative to this value. In the four experiments, the microinjected cells had a GLUT4 signal of 0.99 ± 0.14 (mean ± S.E.). Therefore, GST-SNAP23 did not affect the basal amount of GLUT4 present at the plasma membrane. In insulin-stimulated cells, quantitation of four independent experiments showed that insulin caused a 2.53 ± 0.07 fold (mean ± S.E.) increase in GLUT4 over basal values in membrane lawns from non-injected cells. In vicinal lawns from cells microinjected with GST-SNAP23, the insulin response raised to 3.20 ± 0.30 fold (mean ± S.E.) (Fig. 1). This gain of 26% in insulin response caused by GST-SNAP23 was statistically significant versus non-injected and GST-injected cells (p < 0.05, ANOVA) (Fig. 3b).
To determine if the changes in insulin-stimulated GLUT4 translocation observed using GST-SNAP23 actually resulted in functional GLUT4 exposure at the cell surface we measured glucose uptake in permeabilized cells exposed to GST or GST-SNAP23. Using limited SLO permeabilization of 3T3-L1 adipocytes, a reduced but reproducible insulin-dependent stimulation of glucose uptake is observed, allowing for the introduction of macromolecules to study their effect on insulin action. GST-SNAP23 was introduced into SLO-permeabilized 3T3-L1 adipocytes for 15 min prior to addition of insulin for another 15 min. Glucose uptake was immediately measured as described in Materials and Methods.
Table I Effects of SNAP23 protein on glucose uptake.
Additions Relative glucose uptake
Basal Insulin/basal
None 1.00 ± 0.04 1.50 ± 0.04
GST-SNAP23 0.98 ± 0.19 1.98 ± 0.13*
GST 1.02 ± 0.17 1.52 ± 0.14
Table I shows that introduction of the fusion protein caused a significant enhancement of insulin-stimulated glucose uptake relative to cells exposed to GST or buffer alone. This supports the results in Figure 1 showing that microinjection of GST-SNAP23 increases the level of GLUT4 on membrane lawns. In addition, it demonstrates that this GLUT4 is functionally incorporated into the plasma membrane lawns.
Discussion and Conclusion
The SNARE hypothesis of synaptic vesicle exocytosis initially suggested that SNAP25 is a t-SNARE required for docking/fusion of incoming vesicles. Along with VAMP-2 and syntaxin-1, they constitute the minimum number of proteins that can lead to membrane fusion (11). Subsets of mammalian and yeast SNAREs form very stable ternary complexes via coiled-coil interactions and the energy released from this tight binding is thought to drive membrane fusion (11, 12, 13, 14). In contrast to VAMP-2 and syntaxin-1, which are transmembrane proteins, SNAP25 does not penetrate the membrane but associates with it through palmitoyl residues attached to a set of four cysteine residues in the middle of the molecule (15), and by a secondary mechanism, through coiled-coil interactions with syntaxin-1.
In both 3T3-L1 adipocytes and rat fat cells, GLUT4-containing organelles incorporate into the plasma membrane in response to insulin. This phenomenon has been shown to be sensitive to hydrolysis of VAMP-2 and/or VAMP-3/cellubrevin with botulinum toxins (1, 2), and to neutralization of syntaxin-4 with specific antibodies (3, 16). The results presented here demonstrate that SNAP23 has the ability to bind both syntaxin-4 and VAMP-2 specifically and with high affinity. Any complex formed from all three of these proteins does not appear to be as stable as the analogous complex formed from the neuronal subset of SNAREs as measured by SDS-resistance. This discrepancy with the neuronal complex is not surprising given that the neuronal complex needs to stay in a high energy state until calcium triggers fusion whereas complexes in adipocytes do not appear to be as tightly regulated.
Similar to previous observations made with antibodies directed to syntaxin-4, microinjection of antibodies directed to the C-terminal domain of SNAP23 reduced the insulin-dependent arrival of GLUT4 at the cell surface. This is complementary to a recent report from Rea et al. (17) who used antibodies directed to the N-terminus domain of SNAP23 to show that they also blocked GLUT4 translocation in response to insulin. The fact that both the C-terminus and the N-terminus of SNAP23 appear to be involved in the incorporation of GLUT4 into the membrane is consistent with recent reports that both the N-terminus and the C-terminus of SNAP25 are part of the interacting core of the SNARE complex (18). Furthermore, microinjection of full length SNAP23 into 3T3-L1 adipocytes enhanced the amount of GLUT4 present at the cell surface upon an insulin challenge, while introduction of this protein by chemical permeabilization of the cells allowed for a higher insulin-dependent stimulation of glucose uptake.
The ability of exogenous SNAP23 to enhance the effect of insulin is in contrast to the inhibitory effects of the cytoplasmic domains of syntaxin-4 or VAMP-2, which presumably act as competitive inhibitors of the endogenous, membrane-bound syntaxin-4 and VAMP-2. In those studies both molecules were missing their transmembrane domains, suggesting that this link with the appropriate membranes is critical for their function in incorporating GLUT4 compartments into the plasma membrane. In contrast, the full length SNAP23 is thought to be membrane associated via cysteine palmitoylation and protein-protein interactions, by analogy to SNAP25. We hypothesize that the microinjected SNAP23 protein assists in incorporating the GLUT4 organelle into the plasma membrane by binding to its natural partners syntaxin-4 and VAMP-2. The exogenous SNAP23 would act additively to the (limiting) amount of the endogenous SNAP23.
SNAP23 clearly has the ability to bind syntaxin-4 and VAMP-2, two SNAREs already implicated in insulin-stimulated GLUT4 translocation. Our microinjection and glucose uptake results, combined with those of Rea et al. (17) suggest that SNAP23 may indeed contribute with regions analogous to those of SNAP25, in its interaction with other proteins leading to GLUT4 vesicle docking/fusion in response to insulin. Further, our results show that SNAP23 is important for the functional incorporation of GLUT4 leading to an increase in glucose transport activity.
References
1. B. Cheatham, et al., Proc. Natl. Acad. Sci., USA 93, 15169-15173 (1996).
2. Y. Tamori, et al., Biochem. Biophys. Res. Commun. 220, 740-745 (1996).
3. A. Volchuk, et al., Mol. Biol. Cell 7, 1075-1082 (1996).
4. A. L. Olson, J. B. Knight, J. E. Pessin, Mol. Cell. Biol. 17, 2425-2435 (1997).
5. A. Volchuk, et al., J. Biol. Chem. 270, 8233-8240 (1995).
6. P. P. C. Wong, et al., Biochem. Biophys. Res. Commun. 230, 64-68 (1997).
7. L. J. Foster, et al., Biochemistry 37, 11089-11096 (1998).
8. S.-C. Hsu, et al., Neuron 17, 1209-1219 (1996).
9. A. Volchuk, et al., J. Biol. Chem. 273, 8169-8176 (1998).
10. T. Hayashi, et al., EMBO J. 13, 5051-5061 (1994).
11. T. Weber, et al., Cell 92, 759-772 (1998).
12. P. I. Hanson, R. Roth, H. Morisaki, R. Jahn, J. E. Heuser, Cell 90, 523-535 (1997).
13. D. Fasshauer, H. Otto, W. K. Eliason, R. Jahn, A. T. Brünger, J. Biol. Chem. 272, 28036-28041 (1997).
14. R. C. Lin, R. H. Scheller, Neuron 19, 1087-1094 (1997).
15. M. Veit, T. H. Söllner, J. E. Rothman, FEBS Lett. 385, 119-123 (1996).
16. J. T. Tellam, et al., J. Biol. Chem. 272, 6179-6186 (1997).
17. S. Rea, et al., J. Biol. Chem. 273, 18784-18792 (1998).
18. R. B. Sutton, D. Fasshauer, R. Jahn, A. T. Brunger, Nature 395, 347-353 (1998).
| Discussion Board | Previous Page | Your Symposium |