Poster
Contents
| INABIS '98 Home Page | Your Poster Session | Related Symposia & Posters | Scientific Program | Exhibitors' Foyer | Personal Itinerary | New Search |
Introduction
The CCAAT/enhancer-binding protein alpha (C/EBP alpha) belongs to
a bZIP protein family of transcription factors (1). The bZIP proteins
are characterized by a basic amino acid-rich DNA-binding region and a leucine
zipper domain that is necessary for dimer formation (2). C/EBP alpha conforms
functional homo- or heterodimers with another member of the C/EBP family
and binds the DNA site (TT/GNNGC/TAAT/G) (3,4). This C/EBP alpha binding
site exists in the promoter of some liver-enriched genes, e.g. albumin
and aldolase B (4). Hence, this transcription factor plays important roles
for expression of proteins related to liver functions.
C/EBP alpha is detected at high level in liver, white and brown adipocyte
tissue,placenta, intestine, lung and myeloid cells (5,6,7,8). On the
other hand, it is well known that the C/EBP alpha mRNA level is found abundantly
in differentiated hepatoma cells, but scarcely in dedifferentiated hepatoma
cells (9,10). Moreover primary culture of hepatocytes on collagen-coated
dishes leads to a gradual decrease in C/EBP alpha protein level and DNA-binding
activity. The addition of EHS (Engelbreth-Holm-Swarm) extra cellular matrix
gel in the culture induces re-differentiation of the cultured hepatocytes
and re-expression of C/EBP alpha, as well as remodeling the hepatocyte
morphology and phenotype (11).
Thus C/EBP alpha was closely associated with hepatocyte differentiation
and functions. However the regulation of C/EBP alpha gene expression is
little known in differentiated hepatoma cells. In this study, we cloned
and sequenced 5’-flanking region to elucidate the regulation of the rat
C/EBP alpha gene.
Materials and Methods
(1) Isolation and sequencing of the rat C/EBP alpha gene
A rat genomic library (105 colonies in cosmid vector) was screened
with a rat C/EBP alpha cDNA as describe elswhere. Positive clones were
further analyzed by Southern blot hybridization using the C/EBP alpha cDNA
as a probe. 7.9-kilobase HindIII fragment was subcloned into the Bluescript
II KS (-) vector, and used to generate a series of deletion constructs
corresponding to various restriction enzyme sites. Sequencing of these
subclones was performed by using a Thermo Sequenase fluorescent labelled
primer cycle sequencing kit with 7-deaza-dGTP (Amersham) on the automated
sequencer.
(2) Northern blot analysis
Total RNA from rat liver or cultured cells was extracted using
the guanidium thiocyanate/cesium chloride method. 10ug of total RNA was
electrophoresed through a 1% agarose-6.7% formaldehyde gel and transferred
to nylon membrane. The blots were hybridized according to previous report.
The probe, a 1500bp NruI/NspI fragment from the HindIII-subclone, was labeled
[α-32P]dCTP by the random primer method (TaKaRa).
Results
(1) Cloning and sequence analysis of 5’-upstream region of the rat
C/EBP alpha gene
Previously, we showed decrease in C/EBP alpha gene expression
associated with dedifferentiation of hepatoma cells. To study transcriptional
regulation for the C/EBP alpha gene, we first isolated the rat C/EBP alpha
gene from a genomic library using a 1.5-kilobase cDNA for rat C/EBP alpha
as a probe. Three colonies were identified by the presence of a 7.9-kilobase
HindIII fragment that contained the entire C/EBP alpha gene. And the complete
sequence was determined in approximately 3-kilobase of the 5’-upstream
region. As shown in Fig. 1, the putative binding sites for transcription
factors were found in the 5’-flanking region of the C/EBP alpha gene.
Binding sites for Sp1 and zinc finger protein are found in proximal upstream
of the TATA box. The nucleotide sequence spanning from -195 to -173bp was
well corresponded to the C/EBP binding site in the mouse promoter. The
rat C/EBP alpha promoter possesses a CUP/AP-2 binding site, as well as
the mouse and human promoters. A GT-repeat was mapped at 945bp upstream
from the transcription start site, and C/EBP beta, HNF-3 beta and MyoD
binding sites were also found at distal sites in the 5’-flanking region
of the rat C/EBP alpha gene.
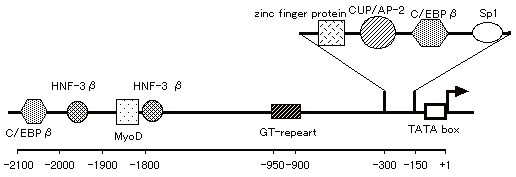 |
Fig. 1 Schematic representation of the DNA: protein interactions in the rat C/EBP alpha promoter. |
(2) Expression of C/EBP alpha mRNA in differentiated and dedifferentiated
hepatoma cells
To study whether the expression of C/EBP alpha mRNA is related
to the differentiation of hepatoma cell lines, we examined the level of
mRNA coding C/EBP alpha in different hepatoma cells. The pattern of C/EBP
alpha mRNA expression in hepatoma cells is shown in Fig. 2. C/EBP alpha
transcript was observed in normal rat liver. Further more, it was detected
in the fully differentiated Reuber cell line at a significant level, while
scarcely in the dedifferentiated variant AH66. Thus the pattern of C/EBP
alpha expression suggested to correlation with the differentiation state
of hepatoma cell lines.
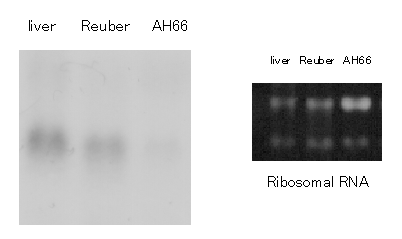 |
Fig. 2 C/EBP alpha mRNA level in differentiated and dedifferentiated
hepatoma cells. Northern blot analysis of the indicated cell line was performed as described in Materials and Methods. |
Discussion and Conclusion
C/EBP alpha has been suggested to play an important role as a key
factor in the switching of proliferation and differentiation, mainly based
on work in adipocyte- and hepatocyte- derived cell lines (11, 12). Recent
study has demonstrated that the transduction of CUP/AP-2 in 3T3-L1 preadipocyte
trans-inhibits the C/EBP alpha gene expression (13). However, the factor
regulating the C/EBP alpha gene is little known in the hepatocyte during
its differentiation. In this report, we have isolated and sequenced the
5’-flanking region of the rat C/EBP alpha gene. We found putative binding
sites for C/EBP beta, HNF-3 beta and MyoD in the upstream region of the
gene. Rana et al. reported previously that C/EBP alpha was transactivated
by C/EBP beta that bound at -190 to -170 in C/EBP alpha gene (14). However,
C/EBP beta binding site of the rat C/EBP alpha gene was additionally located
in distal region. HNF-3 beta is one of the liver-enriched transcription
factors, and first appears in liver development (15). Therefore HNF-3 beta
seems to be associated with the regulation of C/EBP alpha expression in
the hepatocyte differentiation. MyoD, which induces the differentiation
of muscle cells (16), may participate with the down-regulation of the C/EBP
alpha gene.
The expression of C/EBP alpha gene has been shown to be downregulated
in hepatoma cells. The phenotypic difference between Reuber and AH66 seems
to be caused by the amount of C/EBP alpha gene expression. To understand
the correlation between the C/EBP alpha expression and the dedifferentiation
of hepatoma cells, the fine regulatory mechanism of expression C/EBP alpha
gene is necessary further study using differentiated and dedifferentiated
hepatoma cells.
References
- Vinson, C. R., Sigler, P. B. and McKnight, S. L. (1989) Science 246: 911-916.
- Landschulz, W. H., Johnson, P. F. and McKnight, S. L. (1988) Science 240: 1759-1764.
- Cao, Z., Umek, R. M. and McKnight, S. L. (1991) Genes Dev. 5: 1538-1552.
- McKnight, S. L., Lane, M. D. and Gluecksohn-Waelsch, S. (1989) Genes Dev. 3: 2021-2024.
- Xanthopoulos, K. G., Mirkovitch, J., Decker, T., Kuo, C. F. and Darnell Jr., J. E. (1989) Proc. Natl. Acad. Sci. USA 86:4117-4121.
- Williams, S. C., Cantwell, C. A. and Johnson, P. F. (1991) Genes Dev. 5: 1553-1567.
- Birkenmeier, E. H., Gwynn, B., Howard, S., Jerry, J., Gordon, J. I., Landschulz, W. H. and McKnight, S. L. (1989) Genes Dev. 3: 1146-1156.
- Scott, L. M., Civin, C. I., Routh, P. and Friedman, A. D. (1992) Blood 80: 1725-1735.
- Cereghini, S., Yaniv, M. and Cortese, R. (1990) EMBO J. 9: 2257-2263.
- Herbst, R. S., Nielsch, U., Sladek, F., Lai, E., Babiss, L. E. and Darnell Jr, J. E. (1991) New Biol. 3: 289-296.
- Runge, D. Runge, D. M., Bowen, W. C., Locker, J. and Michalopoulos, G. K. (1997) Biol. Chem. 378: 873-881.
- Umek, R. M., Friedman, A. D. and McKnight, S. L. (1991) Science 251: 288-292.
- Jiang, M-S., Tang, Q-Q., McLenithan, J., Geiman, D., Shillinglow, W., Henzel, W. and Lane, M. D. (1998) Proc. Natl. Acad. Sci. USA 95: 3467-3471.
- Rana, B., Xie, Y., Mischoulon, D., Bucher, N. L. and Farmer, S. R. (1995) J. Biol. Chem. 270: 18123-18132.
- Cereghini, S. (1996) FASEB J. 10: 267-282.
- Weintraub, H. (1993) Cell 75: 1241-1244.
| Discussion Board | Previous Page | Your Poster Session |