Invited Symposium: Molecular Mechanisms of Ageing
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Materials and Methods
Materials and Methods Synaptoneurosome Preparation Synaptoneurosomes were isolated from the cortex of 12 day-old Long-Evans rats by sequential syringe filtration through nylon membranes of decreasing pore size followed by low speed centrifugation 1. RNA was prepared using TRI-ReagentTM according to the manufacturer1s directions. Contaminating genomic DNA was removed by precipitation LiCl. Differential Display DDRT-PCR was carried out according the procedure of Sung and Denman to minimize false positive clones 2. One microgram of RNA was converted to first strand cDNA by mixing 10 ml RNA with 2.5 ml of 20 mM of one of three anchored 31 PCR primers (Table 1.) and 9.5 ml of DEPC-H2O. Subsequently, a master mixture consisting of 5 ml 5X reverse transcriptase buffer, 1 ml (40 Units) of RNAsin, 2.5 ml of 0.1 mM DTT, 2.5 ml of 0.25 mM dNTPs and 1 ml (200 Units) of reverse transcriptase was added; the sample was then incubated for 70 min. at 37oC. To produce the differential display, one microliter of each reverse transcription mixture was amplified in the presence of [a-32P] dCTP. Each amplification sample consisted of the following components: 2 ml of 10X Taq DNA polymerase buffer, 1.5 ml of 0.1 mM dNTPs, 1.25 ml (20 mM) of one of six 51 primers (Table 1.), 1 ml of a one-to-five dilution of [a-32P] dCTP (3000 Ci/mmol), 0.5 ml of Taq DNA polymerase and 12.75 ml of DEPC-H2O. The reaction mixtures were amplified using a 25 cycle paradigm consisting of a 94oC, 1.5 min. denaturing step, 42oC, 30 sec. and 57oC, 1 min. annealing steps, and a 72oC, 1.5 min. extension step. Following the reaction, 5 ml of 90% formamide, 20 mM EDTA, 0.3% xylene cyanole and 0.3% bromphenol blue was added; each sample was then boiled for 3 min. Five microliters of the mixture was loaded on to 6% polyacrylamide 7M urea sequencing gels and electrophoresed for 2-3 hr at 1500 V. Subsequently, the gels were transferred to Whatman 3M chromatography paper and dried; differentially displayed bands were detected on Kodak XAR5 film after 1-5 hr of exposure.
Cloning and Sequence Analysis
DDRT-PCR bands were carefully excised from dried gels, rehydrated, reamplified and cloned into pCR-3.1 (Invitrogen). Plasmid DNA, isolated from each of the clones was subjected to double-stranded DNA sequencing in both forward and reverse direction using a 21 base T7 promoter primer and a 21 base M13 reverse primer, respectively. Sequences were checked to insure they included both 51 and 31 sequencing primers; clones fulfilling these criteria for correctness were then compared to GENBank and EST databases with the BLAST (Basic Local Alignment Search Tool) search algorithm, tblastn 3-5, at the National Center for Biotechnology to determine whether the mRNAs had been previously identified. Clone sequences that were not found in the GENBank or EST databases were then queried against the TIGR database to search for tentative consensus sequences. Sequence alignments were performed using the Clustal Multiple Sequence Alignment program 6-8 at the Institute for Biomedical Computing, Washington University, St. Louis.
Touchdown RT-PCR
Touchdown RT-PCR was used to maximize the yield of specific products. Five micrograms of synaptoneurosome and cortex RNAs were first converted to ss cDNA. Ten picomoles of oligo dT was annealed to each RNA in a total of 12 ml for 10 min. at 70oC and then snap-cooled on ice. Subsequently, 4 ml of 5X first strand buffer, 2 ml of 0.1 M dTT and 1 ml of 10 mM dNTPs were added. The contents were mixed and incubated at 42oC for 2 min. One microliter (200U) of Superscript II reverse transcriptase was then added and the incubation was continued for an additional 50 min. Superscript II was heat inactivated at 70oC for 15 min.; RNA was removed with 2 units of RNase H, 37oC, 20 min.
The resulting cDNA was amplified with 5 pmol of each primer, 1.25 U of Taq DNA polymerase and 5 nmol of each dNTP in 25 ml using the following cycling paradigm:
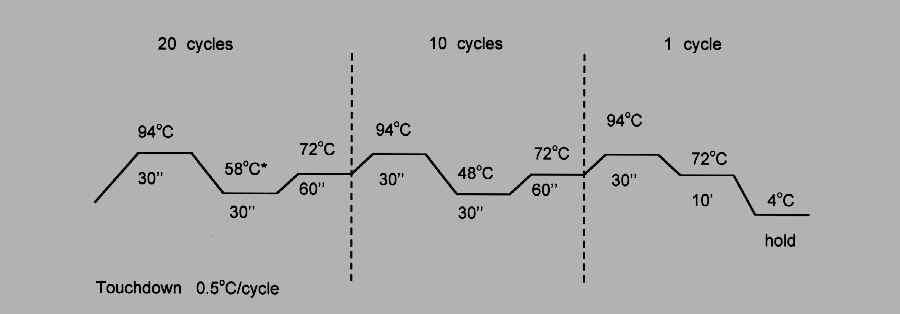 Fig. 1: Touchdown RT-PCR.
Fig. 1: Touchdown RT-PCR.
Various concentrations of cDNA were assessed to insure the amplification of each amplicon was in the linear range.
| <= Introduction | MATERIALS & METHODS | Results => |
| Discussion Board | Next Page | Your Symposium |