Invited Symposium: Molecular Mechanisms of Ageing
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Results
Results To begin to identify mRNAs that are selectively translated at synapses we isolated synaptoneurosomes from the cortex of 12 day-old Long-Evans rats.
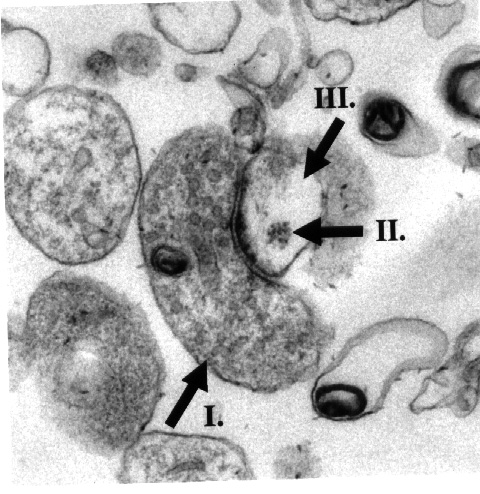 Fig. 2: Electron micrograph of an isolated synpatoneurosome showing a presynaptic terminal (I), a postsynaptic terminal (II) and a polyribosome aggregate (III).
Fig. 2: Electron micrograph of an isolated synpatoneurosome showing a presynaptic terminal (I), a postsynaptic terminal (II) and a polyribosome aggregate (III).
Total RNA isolated from these synaptoneurosomes was compared to total cortex RNA using differential display (DDRT-PCR) with the following set of primers:
Table 1. 3' Primers Sequence _____________________________________________ dT 51 TTT TTT TTT TTT T 31 dT1 51 GCG CAA GCT12 GC 31 dT2 51 GCG CAA GCT12 GG 31 dT3 51 GCG CAA GCT12 GA 31 5' Primers Sequence _______________________________________________ A1 51 CGG ATC GAC TCC AAG 31 A2 51 CGG TAT CGA TAC AGG 31 A3 51 CGG CTG ATC CAT GC 31 A5 51 CGG GTC TGC TAG GTA 31 A6 51 CGG AGA CGC TAG TGT 31 A7 51 CGG ATC TTT CTA CCC 31 _______________________________________________A selection of differentially displayed is shown in Figure 3.
 Click to enlarge
Fig. 3: Representative DDRT-PCR of Synaptoneurosomal (S) and Cortex (S) RNAs. DDRT-PCR bands using three different primer combinations (A) A1/dT1, (B) A2/dT1, (C) A2/dT2. The left pair of DDRT-PCR bands on each gel panel are from reactions with MMLV reverse transcriptase; the right pair or bands are from reactions with and Superscript II reverse transcriptase. Bona fide candidate bands 1-5 (Table 2.) are those bands that are differentially expressed in both sets of RT reactions 9,10. * bands were differentially expressed, but could not be re-amplified.
Click to enlarge
Fig. 3: Representative DDRT-PCR of Synaptoneurosomal (S) and Cortex (S) RNAs. DDRT-PCR bands using three different primer combinations (A) A1/dT1, (B) A2/dT1, (C) A2/dT2. The left pair of DDRT-PCR bands on each gel panel are from reactions with MMLV reverse transcriptase; the right pair or bands are from reactions with and Superscript II reverse transcriptase. Bona fide candidate bands 1-5 (Table 2.) are those bands that are differentially expressed in both sets of RT reactions 9,10. * bands were differentially expressed, but could not be re-amplified.
A total of 60 such bands were isolated using this procedure, 20 of which were cloned and sequenced. A summary of the different clones that were isolated is given in Table 2.
Table 2. Primer Pair1Size Identity _________________________________________________ Actin 246 Ax-actin 1-1 246 Tip60a 1-1 138 Osteonectin 1-2 395 EST193443 1-2 231 Ceramide gluc. transferase 2-2 388 ZBP-89 3-1 155 rpL18a 1-5 95 calmodulin binding protein 2-5 143 EST UI-R-E0-cj-d-08-0-UI.S1 2-5 140 EST-UI-R-E0-bw-f-06-UI.S1 2-5 118 Mitochondrial cytochrome B 2-6 140 Unique 2-7 220 Unique 2-7 197 EST HUM404H10B 2-7 116 rpS29 _________________________________________________ 1. Primer Pair= 31dTx/51Ay where x and y are the different primers shown in Table 1.Touchdown RT-PCR was then used to quantify the message levels in synaptoneurosome RNA and cortex RNA; this allowed us to determine whether the cloned cDNAs were, in fact, derived from mRNAs enriched in synpatoneurosomes. The strategy used is outlined in Figure 4.
 Click to enlarge
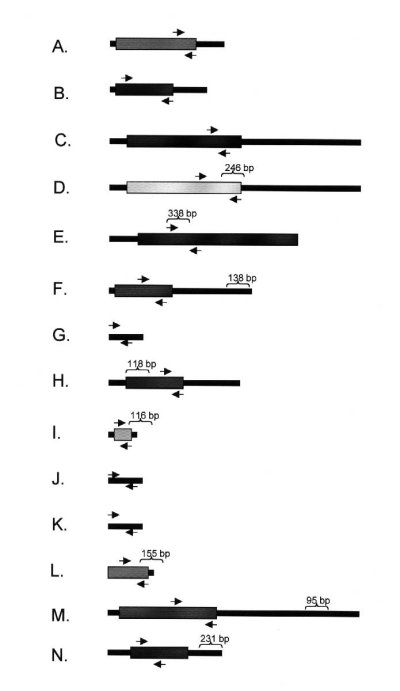
Fig. 4: Primer Design for Touchdown RT-PCR. RT-PCR primers were designed to the coding sequences (CDS) of the eight known mRNAs in Table 2 as well as FMR1 and G3PDH mRNAs that served as controls. Nested primers were designed to three of the four ESTs. Arrows show the location of the forward and reverse primers for each RNA (drawn to scale). Brackets show the location and size of the corresponding DDRT-PCR clone. RNAs for: (A) actin, (B) G3PDH, (C) FMR1, (D) Tip60a, (E) ZBP89, (F) Osteonectin, (G) EST193443, (H) ctyochrome B, (I) ribosomal protein S29, (J) EST UI-R-E0-cj-d-08-0-UI.S1, (K) EST UI-R-EO-bw-f-06-0-UI.S1, (L) ribosomal protein L18a, (M) Vesicle-associated calmodulin binding protein, and (N) ceramide glucosyltransferase were analyzed.
Click to enlarge
Fig. 4: Primer Design for Touchdown RT-PCR. RT-PCR primers were designed to the coding sequences (CDS) of the eight known mRNAs in Table 2 as well as FMR1 and G3PDH mRNAs that served as controls. Nested primers were designed to three of the four ESTs. Arrows show the location of the forward and reverse primers for each RNA (drawn to scale). Brackets show the location and size of the corresponding DDRT-PCR clone. RNAs for: (A) actin, (B) G3PDH, (C) FMR1, (D) Tip60a, (E) ZBP89, (F) Osteonectin, (G) EST193443, (H) ctyochrome B, (I) ribosomal protein S29, (J) EST UI-R-E0-cj-d-08-0-UI.S1, (K) EST UI-R-EO-bw-f-06-0-UI.S1, (L) ribosomal protein L18a, (M) Vesicle-associated calmodulin binding protein, and (N) ceramide glucosyltransferase were analyzed.
Three independent synaptoneurosome/cortex RNA pairs were used in this analysis. The results are summarized in Figure 5.
 Click to enlarge
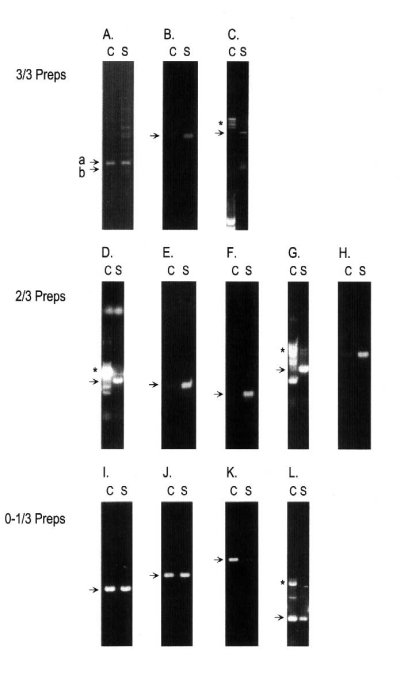
Figure 5. Touchdown RT-PCR of Synaptoneurosome enriched RNAs.
(A-C) RNAs selectively enriched in all three synaptoneurosome (S)/cortex (C) preparations. Panels are (A) Ax-actin (b); and b-actin (a) shown for comparison, (B) ceramide glucosyl transferase, (C) ZBP89.
(D-H) RNAs selectively enriched in two of three three synaptoneurosome (S)/cortex (C) preparations. Panels are (D) vesicle-associated calmodulin binding protein, (E) ribosomal protein L18a, (F) ribosomal protein S29, (G) osteonectin, (H) EST UI-R-E0-cj-d-08-0-UI.S1.
(I-L) RNAs selectively enriched RNAs selectively enriched in only the DDRT-PCR synaptoneurosome (S)/cortex (C) preparation. Panels are (I) EST UI-R-EO-bw-f-06-0-UI.S1, (J) cytochrome c, (K) G3PDH, (L) EST193443. Two amplicons, FMR1 and Tip60a, did not produce specific fragments in any of the RT-PCR assays. * indicates non-specific amplification.
Click to enlarge
Figure 5. Touchdown RT-PCR of Synaptoneurosome enriched RNAs.
(A-C) RNAs selectively enriched in all three synaptoneurosome (S)/cortex (C) preparations. Panels are (A) Ax-actin (b); and b-actin (a) shown for comparison, (B) ceramide glucosyl transferase, (C) ZBP89.
(D-H) RNAs selectively enriched in two of three three synaptoneurosome (S)/cortex (C) preparations. Panels are (D) vesicle-associated calmodulin binding protein, (E) ribosomal protein L18a, (F) ribosomal protein S29, (G) osteonectin, (H) EST UI-R-E0-cj-d-08-0-UI.S1.
(I-L) RNAs selectively enriched RNAs selectively enriched in only the DDRT-PCR synaptoneurosome (S)/cortex (C) preparation. Panels are (I) EST UI-R-EO-bw-f-06-0-UI.S1, (J) cytochrome c, (K) G3PDH, (L) EST193443. Two amplicons, FMR1 and Tip60a, did not produce specific fragments in any of the RT-PCR assays. * indicates non-specific amplification.
| <= Materials & Methods | RESULTS | Discussion & Conclussions => |
| Discussion Board | Next Page | Your Symposium |