Invited Symposium: Medicinal Plants and Drug Actions
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Introduction
Ginseng is one of the most highly valued Chinese herbal medicines and the three important species are P. ginseng, P. quinquefolius, and P. notoginseng. P. ginseng is known as ginseng; P. quinquefolius is known as American ginseng; and P. notoginseng is known as San Chi. P. ginseng and, P. quinquefolius are very similar in morphology and they both are called ginseng, but they are actually grown in two separated continents (Asia and American) (Wen and Zimmer, 1996). They have diverse and sometime opposite physiological activities based on the different combination and the quantity of various ginsenosides found in each species (Kwan, 1995).
Traditionally, authentication of the P. ginseng, P. quinquefolius, and P. notoginseng is based on morphological characteristics. Recently, new biochemical analysis of the ginsenosides by high pressure chromatography (HPLC) and, at the molecular level, arbitrarily-primed polymerase chain reaction (AP-PCR) and random amplified polymorphic DNA have been used to differentiate the three medicinal species of panax from one another and also from their common adulterants. (Shaw and But, 1995).
DNA fingerprinting assay is a molecular technique for detecting genetic differences at a large number of hypervariable DNA loci first introduced by Jeffreys et al (1985a). These hypervariable minisatellite regions, often called variable number of tandem repeat (VNTR) consist of tandem repeat of short segment of DNA and alleles arise from variation in the number of repeats (Jeffreys 1987, Nakamura et al. 1987). When these repeats are labeled and used as a probe in DNA fingerprinting assay, individual-specific DNA profile can be obtained providing a powerful application to human forensics (Gill et al. 1985, Jeffreys et al. 1985b, Jeffreys et al. 1985c). Since repetitive DNA is a major component of the plant genome and may account for more than half of the genome, the application of VNTR DNA markers in plants is increasing (Avise 1994, Weising et al. 1995). Rogstad (1996) used VNTR DNA markers to detect levels of outcropping in plants, distinguishing between selfing and apomixis, and demonstration of agamospermy in tarazacum. Since studies related to the molecular genetics of Chinese medicinal plant species are limited, and one of the limitations in DNA fingerprinting ginseng has been the lack of or limited availability of VNTR probes. We have adopted a novel approach named as low-Cot DNA probe method. This low-Cot DNA probe was first developed for the identification of host-specific DNA fragments in Fusarium oxysporum and it was found to be an excellent DNA probe for DNA fingerprinting (Leung et al. 1992).
This method was later used in developing DNA fingerprinting probes for salmon (Leung et al. 1994) and Amaranthus (Sun et al. in preparation). Microsatellite consists of short tandem repetitive DNA of less than 6 bp, and it has been demonstrated to be highly informative. It is abundant and evenly distributed in the plant genome (Wang et al., 1994; Lagercrantz et al. 1993; Smith and Devey, 1994; Depeiges et al. 1995), specially the (AT) n and (A/T) n (Wang et al, 1994; Depeiges et al. 1995), In this paper, we discuss the use of the microsatellite and low-Cot DNA as a DNA probe for fingerprinting P. ginseng and P. quinquefolius.
Materials and Methods
Plant materials
The plants materials used in this study were obtained either from local market or purchased from the Institute of Medicinal Plant Development in Beijing. Fresh/dried root of P. ginseng and dried root of P. quinquefolius were purchased from the Institute of Medicinal Plant Development, Peking Union Medical College, Chinese Academy of Medical Sciences. Other plant materials such as carrot, celery, parsley, pea and potato were purchased from local supermarkets. Dried plant materials were stored at room temperature in airtight containers while fresh materials were kept frozen at -200C.
DNA extraction
All plant genomic DNA was extracted using a modified cetyltrimethyl ammonium bromide (CTAB) protocol (Reichardt and Roger, 1995). The modified method combines a cell lysis step by SDS and a high salt precipitation step for the removal of the polysaccharides in the ginseng samples. Additional washing with TE buffer was added. In brief, two grams of dried or fresh samples were ground in liquid nitrogen into fine powder. The powder was then transferred into a 15-ml conical tube containing 15 ml of pre-warmed extraction buffer (100mM Tris-HCl, pH8, 50mM EDTA, 900mM NaCl, 1% SDS and 2% beta-mercaptoethanol) and mixed thoroughly. After 30 minutes incubation at 650C, 5M NaCl was added to increase the NaCl concentration to 1.4mM and followed with the addition of one fifth volume of 5% CTAB solution and mixed. The mixture was incubated in a 650C water bath for 20 minutes and then mixed with equal volume of chloroform:isoamylalcohol (24:1). After centrifugation at 3,838xG, 40C for 10 minutes, one third volume of CTAB precipitation buffer (1% CATB, 50mM Tris, pH8, 10mM EDTA, pH8) was mixed with the supernatant to lower thew salt concentration and then 0.6 volume of isopropopanol was addded to precipitate the CTAB-DNA complex. Storage in -200C for 30 minutes may be necessary if no precipitate is found immediately.
After centrifugation at 3,838xG, 40C for 10 minutes, the pellet were dried completely in a speedvac. 1 ml of TE buffer was added to resuspend the pellet in 650C with occasional shaking until the pellet disperse and the suspension become turbid. In this washing step, most of the remaining polysaccharide will dissolve in the TE buffer while most of the CTAB-DNA complex remain insoluble in the low salt condition. Centrifugation at 3,838xG, 40C were carried out to collect pellet and 0.5 ml of high salt TE was added to precipitate the DNA. The DNA pellet was collected by centrifugation and was washed in 70% ethanol. Since CTAB is freely soluble in ethanol, most of the CTAB residues will be removed after ethanol wash. DNA was dried in a speedvac and resuspended in an appropriate amount of TE buffer depending on the size of the pellet.
Isolation of low-Cot DNA and cloning of the repetitive sequences
Total genomic DNA from fresh P. ginseng root was used for the isolation of low-Cot DNA according to the method first developed for the identification of pathogenic fungi (Leung et al. 1992). To isolate the low-Cot DNA from P. ginseng, five hundred microgram of total genomic DNA was sheared into smaller than 2kb size fragments by a chemical method described by Britten et al. (1974). The sheared DNA was denatured in boiling water and reannealed for 3 hours at 600C, 0.12M phosphate buffer solution and 0.18M NaCl. The reannealed double-stranded DNA was recoved by hydroxylapatite column chromatography. The yield of the reannealed DNA was measured by fluorometer and the Cot value is calculated as suggested by Britten and Kohne (1968). The isolated DNA fraction is called the low-Cot DNA.
DNA fingerprinting
Six microsatellite probes, including the (CAC)5, (CA)12, (GATA)4, (GACA)4, (GGAT)4, and (GAA)5, and the low-Cot DNA were labeled by digoxigenin (DIG) using the DNA DIG Labeling KitTM (Boehringer Mannheim, Germany) and the PCR DIG Labeling KitTM (Boehringer Mannheim, Germany), respectively, according to the standard protocols provided by the manufacture.
Genomic DNA from individual root was digested overnight with restriction enzymes, and the digested DNA samples were size-fractionated by 1% agarose gel electrophoresis. Vacuum-blot transfer (Pharmacia Biotech, Sweden) was carried out to transfer DNA from agarose gel onto nylon membrane. The hybridization and detection procedure were carried out according to the manufacturer's instructions, and the CDP- StarTM (Boehringer Mannheim, Germany) was used as substrate.
Results
None of the microsatellite probes gave informative fingerprints when used as probes in the DNA fingerprinting assay. Smears of unresolved bands were detected when (CA)12 was used as a probe and the remaining probes generated no DNA fingerprinting signals (data not shown) suggesting low abundance of these microsatellite sequences in the ginseng genome. We used genomic dot blot hybridization to examine the genomic abundance of these plant microsatellite sequences. When the membranes containing serial dilution of genomic DNA from P. ginseng, P. quinquefolius, pea, and potato were hybridized with each of the four microsatellite probes, the hybridization signals detected from P. ginseng and P. quinquefolius were much less than those observed from potato genomic DNA as seen in Figure 1. All six microsatellite probes tested were not suitable to be used as probes, and may be due to the low copy number present in the ginseng genome.
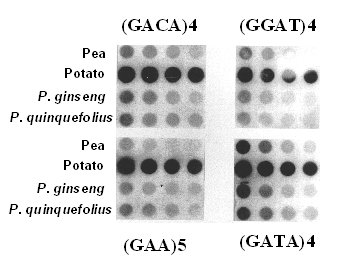 Figure 1. Dot blot analysis of genomic DNA serial diluted from pea, potato, P. ginseng, and P. quinquefolius hybridized with (GACA) 4, (GGAT)4, (GAA)5, and (GATA) 4.
Figure 1. Dot blot analysis of genomic DNA serial diluted from pea, potato, P. ginseng, and P. quinquefolius hybridized with (GACA) 4, (GGAT)4, (GAA)5, and (GATA) 4.
We then isolated the rapid reassociating fraction of the sheared P. ginseng total genomic DNA, called low-Cot DNA. The rapid re-annealing DNA fraction represents approximately 6% of the P. ginseng genome and the calculated Cot value is 1.97. When this fraction of the P. ginseng genomic DNA was randomly labeled with DIG and used as a probe, distinctive fingerprinting profiles were detected among P. ginseng, and P. quinquefolius as seen in Figure 2 and Figure 3. Dar I digested DNA revealed the highest number of resolvable DNA fragments and Taq I showed the lowest number of bands. Combining figure 2 and 3, similarity index (SI) between P. ginseng, and P. quinquefolius is 0.55 and the SI is only 0.06 between pea and P. ginseng and 0.04 between pea and P. quinquefolius, respectively.
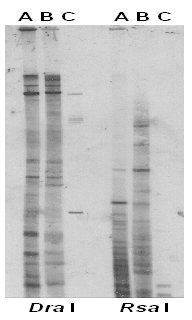 Figure 2. DNA fingerprints of P. ginseng (A), P. quinquefolius (B), and Pea (C), genomic DNA digested with Dra I and Rsa I probed with the P. ginseng low-Cot DNA probe.
Figure 2. DNA fingerprints of P. ginseng (A), P. quinquefolius (B), and Pea (C), genomic DNA digested with Dra I and Rsa I probed with the P. ginseng low-Cot DNA probe.
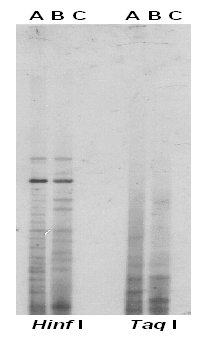 Figure 3. DNA fingerprints of P. ginseng (A), P. quinquefolius (B), and Pea (C), genomic DNA digested with Hinf I and Taq I probed with the P. ginseng low-Cot DNA probe.
Figure 3. DNA fingerprints of P. ginseng (A), P. quinquefolius (B), and Pea (C), genomic DNA digested with Hinf I and Taq I probed with the P. ginseng low-Cot DNA probe.
Discussion and Conclusion
Repetitive sequences are the source for polymorphic markers that are important for DNA fingerprinting probe development and for genome mapping. Traditionally, two widely used cloning strategies for identifying repetitive DNA sequences. One is to enzyme digestion. Genomic DNA is digested with a variety of restriction endonuclease and size fractionated the digested genomic DNA in agarose gel. When distinctive DNA band is detected, the band is excised, subcloned and sequenced. It is usually found to contain a family of repetitive sequences. This method is common and has been used successfully in isolating a number of novel repetitive DNA sequences. The second method is screening genomic library by using known repetitive sequences as a probe. This method is widely used for the isolation of microsatellite (Depeiges et al, 1995, Morchen et al. 1996). Both methods are labor intensive, and time consuming. In contrast, the low-Cot DNA approach that we used allows us to generate a DNA fingerprinting in a very short time. It also allows us to clone for further characterization of the repetitive sequences. Our present study extends the utility of the low Cot DNA probe approaches and confirms its application as a probe in DNA fingerprinting assay.
The possibility of using a microsatellite for fingerprinting Panas species was tested. Our results indicated that some of the most widely used microsatellite DNA fingerprinting probes were not useful for fingerprinting ginseng because of their low copy number or lack of polymorphism. Although the microsatellite probes used in this study were proven to be highly informative in animal and plant, their low abundance in the Panax species limited their use in DNA fingerprinting. Our results do not exclude the possibility that others microsatellite repeats may be informative for fingerprinting ginseng. However, it may involves a large scale screening of all possible combination of short repeats or screening of genomic library.
In summary, we have demonstrated the utility of the low-Cot DNA as a probe in DNA fingerprinting. The high similarity index between P. ginseng and P. quinquefolius is very interesting and it agrees with the low internal transcribed spacers sequence divergence reported by Wen and Zimmer (1996). Therefore, low-Cot DNA probe is a suitable probe to be sued for population study of ginseng, and it should be a useful tool in the breeding and cultivation programs for P. ginseng and P. quinquefolius.
References
- Avise, J.C. 1994. Molecular markers, natural history, and evolution, Chapman & Hall, New York.
- Britten, R.J. and D.E. Kohne, 1968. Repetitive sequences in DNA. Science 161:529-540.
- Britten, R.J., D.E. Graham, and R.Nuefeld, 1974. Analysis of repeating DNA sequences by reassociation. In Nucleic Acidsand Protein Synthesis. Eds L. Grossman and K. Moldave, Academic press, New York, Vol XXIX:363-405.
- Burke, T., D. Dolf, A.J. Jeffreys, and R. Wolff, 1991. DNA fingerprinting: approaches and applications. Birkhauser Verlag, Basel.
- Depeiges, A., Goubely, C., Lenoir, A., Cocherel, S., Picard, G., Raynal, M., Grellet, F. and Delseny, M. 1995. Identification of the most represented repeated motifs in Arabidopsis thaliana microsatellite loci. Theoretical and Applied Genetics 91:160-168.
- Gill, P., A.J. Jeffreys, and D.J. Werett, 1985. Forensic application of DNA "fingerprints" Nature 318:577-579.
- Jeffreys, A.J., 1987. .Highly variable minisatellites and DNA fingerprinting Biochem. Soc. Trans. 15:309-317.
- Jeffreys, A.J., J.F.Y. Brookfield, and R. Semenoff, 1985. Positive identification of an immigration test-case using human DNA fingerprints. Nature 317:818-819.
- Jeffreys, A.J., V. Wilson, and S. L. Thein, 1985a: Hypervariable "minisatellite" regions in human DNA. Nature 314:67-73. Jeffreys, A.J., V. Wilson, and S. L. Thein, 1985b Individual-specific "fingerprints" of human DNA. Nature 316:76-79.
- Kwan, C.Y. 1995. Vascular effects of selected antihypertensive drugs derived from traditional medicinal herbs. Clinical and Experimental Pharmacology and Physiology (Supplement) 1:297-299.
- Langercrantz, U., Ellegren, H., and Andersson, L 1993. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Research 21:1111-1115.
- Leung, F.C., D.P. Chandler, X-Z Shen, A.E. Jarrell, and R.J. Fellow, 1992: Use of repetitive sequences for the identification of host-specific DNA fragments in Fusarium oxysporum. In Americian Phytopathological Society Meeting, Portland, Oregon, abstract A28.
- Leung, F.C., M. Welt, R.D. Quesenberry, and X-Z Shen, 1994 DNA fingerprinting and cloning of hypervariable minisatellite repeats in salmonids. Can. J. Fish. Aquat. Sci. 51(Suppl.1):258-266.
- Martin, E. Fujimoto, M. Hoff, E. Kumlin, and R. White, 1987. Variable number of tandem repeat (VNTR) markers for humne gene mapping. Science 235:1616-1622.
- Morchen, M., Cuguen, J., Michaelis, G., Hanni, C., Saumitou-Laprade, P. 1996. Abundance and length polymorphism of microsatellite repeats in Beta vulgaris. Theoretical and Applied Genetics.
- Reichardt, M, and S. Roger, 1995. Preparation of plant DNA using CTAB, in Current protocol in molecular biology, 2nd ed. Ausubel, F.M., R. Brent, R.E. Kinston, D.D. Moore, J.G. Seidman, J.A. Smith, and K.Struhl (eds). Vol. 1. p. 2.3.3-2.3.7. John Wiley & Sons, Inc, New York.
- Rogstad, S.H. 1996 Assessing genetic diversity in plants with synthetic tandem repetitive DNA probes, in Genomes of plants and animals: 21st stadler Genetics Symposium, Gustafson, J. P., and R.B. Flavell (eds). Plenum Press, New York.
- Shaw, P-C, P. P-H But, 1995. Authentication of Panax species and their adulterants by random-primed polymerase. Planta Med: 61:466-469
- Smith, D.N. and Devey, M.E. 1994. Occurrence and inheritance of microsatellites in Pinus radiata. Genome 37:977-983.
- Wang, Z., Weber, J.L., Zhong, G. and Tanksley, S.D. 1994. Survey of plant short tandem DNA repeats. Theor and Applied Genetics 88:1-6.
- Weising, K., H. Nybom, K. Wolff, and M. Wieland, 1995. DNA fingerprinting in plants and fungi. CRC Press, Boca Raton.
- Wen, J. and E.A. Zimmer 1998 Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol. Phylogenet. Evol. 6:167-177.
| Discussion Board | Previous Page | Your Symposium |