Cell Biology Poster Session
| INABIS '98 Home Page | Your Session | Symposia & Poster Sessions | Plenary Sessions | Exhibitors' Foyer | Personal Itinerary | New Search |
Introduction: cell deaths programmed by selfish genetic elements
Selfish genetic elements are often in conflict with the remainder of the genome1. Prokaryotic prophage (provirus) genomes provide an example. Plasmids and transposons are other examples. Cell death is often programmed to occur under certain conditions by resident selfish genetic elements2. The altruistic suicide of cells infected with viruses serves to counteract secondary infection of neighboring cells (Fig. 1). In the prokaryotes, this type of cell death belongs to the phage exclusion phenomena and is programmed by resident genetic elements, such as prophages and plasmids3.
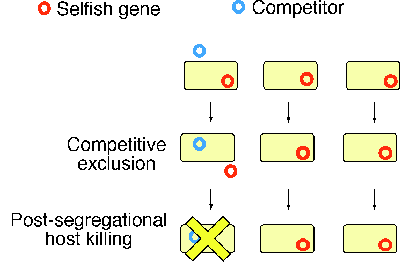 Figure 1
Figure 1
A second class of cell deaths mediated by selfish genetic elements is a plasmid-maintenance mechanism called post-segregational host killing4. A killer and anti-killer gene pair on a plasmid renders the bacterial host “addicted” to the continual presence of this "dispensable" genetic element2. Plasmid loss leads to an imbalance in the action of killer and anti-killer gene products and consequent cell killing through a variety of mechanisms. When a plasmid is displaced by an incompatible competitor plasmid, this host killing would eliminate this competitor together with the host (Fig. 1). The copies of the plasmid would survive in the neighboring clonal cells enjoying resources that would be unavailable without this killing.
In this review, I argue that the restriction modification (RM) gene complexes, such as EcoRI, belong to these selfish genetic elements programming cell deaths5,6,4. I attempt to explain features of RM systems and prokaryotic homologous recombination from this point of view.
Post-segregational cell killing by restriction modification systems
Restriction modification systems and the cellular defense hypothesis
A type II restriction endonuclease (R) makes a double-strand break within, or close to, a specific recognition sequence on duplex DNA (Fig. 1 i). A cognate modification enzyme (M) can methylate the same sequence and protect it from the cleavage7. The tight association of a restriction gene with a cognate modification gene has been termed (type II) restriction modification (RM) gene complex or system. RM gene complexes are abundant in the prokaryotes and in the archaebacteria. Some of them are on plasmids, while others are on the bacterial chromosomes. The genome projects revealed that a bacterial cell can contain multiple RM gene complexes or its homologues8.
Restriction enzymes will cleave foreign DNA, such as viral and plasmid DNA, when this DNA has not been modified by the appropriate modification enzyme (Fig. 2 ii).
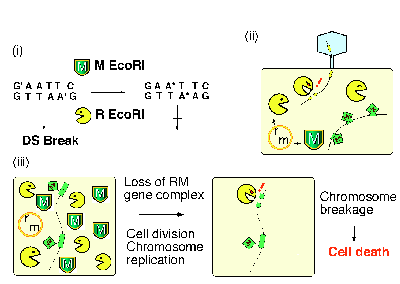 Figure 2
Figure 2
Therefore, it has been widely believed that the evolution and maintenance of RM systems have been driven by the cell's necessity to protect itself against infection by foreign DNA. There are, however, several issues that are not satisfactorily explained by this “cellular defense hypothesis”5.
Post-segregational host killing by restriction modification systems
We found that a plasmid carrying an RM gene complex could not be readily displaced by a second, incompatible plasmid, even when the incoming plasmid was resistant to restriction by the resident RM system9,10. This resistance of RM gene complexes to displacement turned out to be because of the death of cells that have lost the RM gene complexes. Several type II RM gene complexes can increase the apparent stability of a plasmid that carries them by post-segregational host killing in a pure culture9-12, 22.
Our measurements suggested the following course of events after a cell has lost its RM gene complex (Fig. 2 iii). The cell’s descendants will contain fewer and fewer molecules of the modification enzyme. Eventually, the enzyme’s capacity to modify the many sites needed to protect the newly replicated chromosomes from the remaining pool of restriction enzyme will become inadequate. Chromosomal DNA will then be cleaved at the unmodified sites, and the cells will be killed9-11. The selfish-gene hypothesis for restriction modification systems
The host-killing strategy of an RM gene complex makes any DNA that carries the RM gene complex an effective competitor when faced with other DNA that can exclude it (Fig. 1). After host killing, copies of the RM gene complex would survive in the neighboring clonal cells, enjoying resources that would be unavailable if the cells carrying the competitor genetic element had not died (Fig. 1). It is possible that this competitive advantage drove the spreading of RM systems and continues to assure their maintenance. This selfish-gene hypothesis for RM systems9-11,4,5,6 provides explanations for several observations that are difficult to account for by the cellular defense hypothesis.
The selfish-gene concept for the RM gene complexes implies a certain degree of independence from the rest of the genome and consequent movement of RM gene complexes between genomes. There are observations consistent with the mobility of RM gene complexes between bacterial cells and between genomes. Those RM gene complexes on the chromosome will move between bacteria by natural transformation. Those RM gene complexes on plasmids will move between bacteria by conjugation. Some RM complexes are on prophage-like elements13, and some are linked with a site-specific recombinase homologue14 or a transposase homologue15. The EcoRI RM unit appears to have been inserted into a plasmid, while the HaeII RM unit appears to have been inserted into a preexisting operon32. There are examples suggesting horizontal transfer of RM genes between different bacterial genera16,17.
The physical separation of the restriction and modification activities on different proteins contrasts with the tight linkage of their genes. The tight genetic linkage will allow simultaneous loss of R and M genes, while the physical separation of the enzymatic activities will allow execution of host killing by the restriction enzyme. Indeed no post-segregational killing was observed for type I RM systems, in which restriction enzyme activity and modification enzyme activity can be physically associated18, 19.
Competition between restriction modification systems
Sequence recognition
The sequence recognition by the RM systems is individually highly specific and collectively quite diverse. In this respect, RM proteins are unique among DNA binding protein families. The selfish-gene hypothesis explains this by competition among selfish-gene entities for recognition sequences as follows.
If two RM systems present in the same host cell recognize different sequences, displacement of even one of them will lead to killing of the host cell through chromosome cleavage (Fig. 3 middle). Therefore, each of these two RM systems can force its maintenance on the host cell. The host killing by an RM gene complex will not work when the second RM gene complex shares the same sequence specificity. Methylation of the recognition sites along the chromosome by the modification enzyme of one RM will protect them from cleavage by the restriction enzyme of the other RM (Fig. 3 right). Therefore, two RM systems of the same sequence specificity will not be able to enjoy stabilization simultaneously. These predictions were confirmed in experiments11.
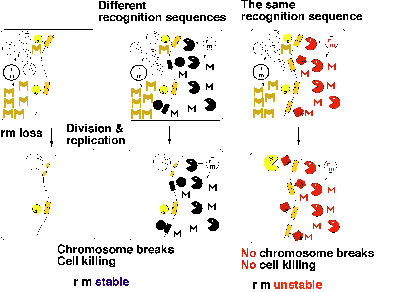 Figure 3
Figure 3
We may call this phenomenon "incompatibility" between RM systems. Each recognition sequence would define one incompatibility group. This type of incompatibility implies competition for specific sequences by RM systems. This would result in the specialization of each of these selfish-gene units in only one of many diverse sequences. This explains the diversity and specificity in their sequence recognition. Mutual exclusion
A second type of competition between two RM gene complexes was identified during analysis of a regulatory gene --- the C gene --- tightly linked with several RM gene complexes20. The C gene is required for restriction. The C regulatory system helps establishment of RM systems in a new host bacterial cell. It ensures that restriction enzyme is made after the M gene has been expressed to modify sufficiently the host chromosome (Fig. 4 i).
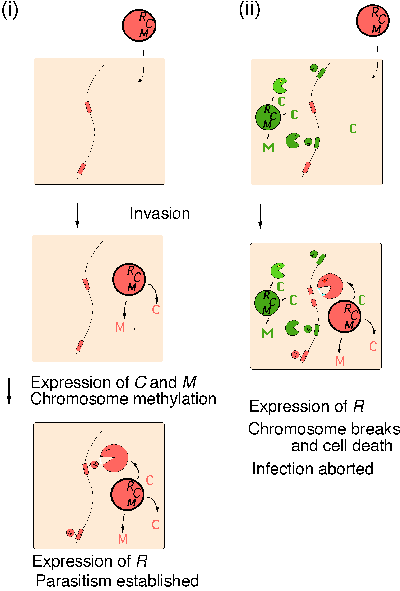 Figure 4
Figure 4
BamHI C protein and PvuII C protein can cross-complement21. Presence of the BamHI RM system in a host cell aborts invasion of the PvuII RM system by eliciting host killing through premature restriction by PvuIIR22 (Fig. 4 ii). The experimental results suggest that the resident BamHI C protein forces the expression of incoming PvuII R gene, which leads to cleavage of yet-unmodified PvuII sites along the chromosome. This novel phenomenon, called “apoptotic mutual exclusion”, is similar to suicidal defense against phage invasion (see Introduction and Fig. 1) and may well play a similar role in a longer time scale.
The exclusion phenomenon defines another level of incompatibility between RM systems. PvuII and BamHI define one incompatibility group, and EcoRV defines another22. This incompatibility operates between RM systems with different recognition sequences and is, therefore, complementary to the incompatibility based on the identity of the recognition sequence. Classification in terms of these two sorts of incompatibility will form a basis for biology of RM systems.
Homologous recombination as adaptation
The selfish-gene hypothesis for RM systems might provide a clue to an important issue --- the forces underlying the evolution and maintenance of sex. Sex is defined here as homologous recombination often involving outcrossing (i.e. pairing of DNAs from different clones) and crossing-over. It is ubiquitous in bacteria and bacteriophages. The previous hypotheses proposed repair of DNA damages, elimination of deleterious mutations and escape from parasites as the likely forces. Two kinds of homologous recombination, one by bacteriophages and the other by bacteria, can be understood as responses to DNA double-strand breaks23. The parasitism of RM systems might have been important in their evolution. Double-strand break repair by bacteriophages
In the double-strand break repair models24,25, a double-strand break is repaired by copying a homologous DNA, and this repair is often accompanied by crossing over of flanking sequences. This reaction starting from type II restriction breaks has been demonstrated for recombination machinery carried by lambdoid bacteriophages26-28. It is conceivable that this double-strand break repair mechanism represents one form of adaptation of bacteriophages to the attack by RM units28 (Fig. 5A).
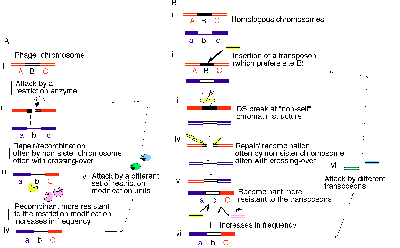 Figure 5
Figure 5
First of all, restriction cleavage of a recognition site in the phage DNA would stimulate its repair with an uncut DNA, usually a phage chromosome that belongs to the same clone.
The effect of the repair on phage clone survival could become larger if it were to take place with a partner from another phage clone possessing a divergent genome. When the template DNA lacks the recognition site, recombination might result in a DNA region devoid of this particular restriction site (Fig. 5A).
The crossing-over of flanking sequences triggered by the break would confer a third kind of advantage. In outcrossing, alleles at different locations would be recombined to generate rare combinatory genotypes. Some of them would be more resistant to the attack by the present RM systems than the currently major combinations, and they would increase in number (Fig. 5A).
As a phage population encounters bacterial populations possessing various combinations of RM systems of diverse specificities, the process of breakage, repair, gene conversion and crossing-over will continue (Fig. 5A). The continual race between hosts (i.e. bacteriophage DNAs) and these genomic parasites (i.e. RM gene complexes) would favor the evolution of sex by the bacteriophages.
I have presented analogous arguments for eukaryotic meiosis and sex as a strategy against parasitic DNAs28, 29, 6 (Fig. 5B). Bacterial homologous recombination
A major route of homologous recombination by E. coli is mediated by a system composed of RecBCD exonuclease, RecA protein, and a specific 8-mer sequence, 5’ GCTGGTGG, called chi, found at high frequency in the E. coli chromosome30,31. The recombination is initiated by a double-strand break on duplex DNA. From the break, RecBCD enzyme starts exonucleolytic DNA degradation. When the enzyme encounters chi, it attenuates degradation and, together with RecA protein, promotes recombination of this DNA with a homologous DNA.
The cell killing following loss of the EcoRI gene complex is counteracted by RecA/RecBCD/chi system (N. Handa, A. Ichige, K. Kusano, I. Kobayashi, unpublished). It repairs the broken chromosomes and allows the cell to revive. This result led to an explanation for the strange destruction/ recombination behavior of the RecABCD system in terms of collaboration and fights among three genetic elements --- RM system, invading DNA, and the host Rec system --- within a bacterial cell28, 33. The host Rec system would destroy invading nonself DNA after restriction cleavage. On the other hand, it would allow recombinational restoration of its own chi-marked chromosomal DNA after its restricting cleavage (Fig. 6). If incoming DNA carries chi in some proper configuration, the exonucleolytic degradation would be attenuated and parts of the fragment may be incorporated into the chromosome --- if not rejected by the mismatch repair system. This would result in the mosaic polymorphisms of the chromosome within a bacterial group34, 35.
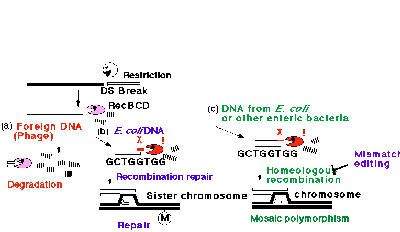 Figure 6
Figure 6
Acknowledgements
I am grateful to my lab members for helpful discussions. The work from our group has been supported by grants from Ministry of ESSC of Japanese government.
References
1 Hurst, L.D., Atlan, A. and Bengtsson, B.O. (1996) Quart. Rev. Biol. 71, 317-364
2 Yarmolinsky, M.B. (1995) Science 267, 836-837
3 Snyder, L. (1995) Mol. Microbiol. 15, 415-420
4 Gerdes, K., Ayora, S., Canosa, I., Ceglowski, P., Diaz, R., Franch, T., Gultyaev, A. P., Jensen, R. B., Kobayashi, I., Macpherson, C., Summers, D., Thomas, C., Zielenkiewicz, U. (in press) in Plasmid biology (Thomas, C. M. editor-in-chief), Harwood academic publishers gmbh, Amsterdam
5 Kobayashi, I. (1996) in Epigenetic mechanisms of gene regulation (Russo, V., Martienssen, R. and Riggs, A., eds), pp. 155-172, Cold Spring Harbor Laboratory Press, New York
6 Kobayashi, I. (1998) Trends in Genetics 14, 368-374
7 Roberts, R.J. and Macelis, D. REBASE: The restriction enzyme database. URL: http://www.neb.com/rebase
8 TIGR Microbial Database: a listing of microbial genomes completed and in progress. URL: http://www.tigr.org/tdb/mdb/mdb.html
9 Naito, T., Kusano, K. and Kobayashi, I. (1995) Science 267, 897-899
10 Naito, Y., Naito, T. and Kobayashi, I. Biol. Chem. 379, 429-436(1998)
11 Kusano, K., Naito, T., Handa, N. and Kobayashi, I. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11095-11099
12 Kulakauskas, S., Lubys, A. and Ehrlich, S.D. (1995) J. Bacteriol. 177, 3451-3453
13 Anton, B.P. et al. (1997) Gene 187, 19-27
14 Vaisvila, R., Vilkaitis, G. and Janulaitis, A. (1995) Gene 157, 81-84
15 Brassard, S., Paquet, H. and Roy, P.H. (1995) Gene 157, 69-72
16 Wilson, G.G. and Murray, N.E. (1991) Annu. Rev. Genet. 25, 585-627
17 Jeltsch, A. and Pingoud, A. (1996) J. Mol. Evol. 42, 91-96
18 O’Neill, M., Chen, A. and Murray, N.E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14596-14601.
19 Kulik, E. M. and Bickle, T. A. (1996) J. Mol. Biol. 264: 891-906.
20 Tao, T., Bourne, J.C. and Blumenthal, R.M. (1991) J. Bacteriol. 173, 1367-75
21 Ives, C. L., Sohail, A. and Brooks, J. E. (1991) J Bacteriol. 177, 6313-6315
22 Nakayama, Y. and Kobayashi, I. Proc. Natl. Acad. Sci. U.S.A. 95, 6442-6447 (1998)
23 Kobayashi, I. (1992) Adv. Biophysics 28, 81-133
24 Resnick, M.A. (1976) J. Theor. Biol. 59, 97-106
25 Szostak, J.W., Orr-Weaver, T.L., Rothstein, R.J. and Stahl, F.W. (1983) Cell 33, 25-33
26 Takahashi, N. and Kobayashi, I. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2790-2794
27 Tomoki Yokochi, T., Kusano, K. and Kobayashi, I. (1995) Genetics 139: 5-17
28 Takahashi, N. K. et al. (1997) Genetics 146, 9-26
29 Kobayashi, I. (in press) Ann. New York Acad. Sci.
30 Kowalczykowski, S.C. et al. (1994) Microbiol. Rev. 58, 401- 465
31 Blattner, F.R. et al. (1997) Science 277, 1453-1474
32 Stein, D. et al. (1998) Biol. Chem. 379, 575-578
33 Handa, N. et al. (1998) Microb. Comp. Genomics 2, 287-298
34 Smith, J.M., Dowson, C.G., Spratt, B.G. (1991)Nature. 349, 29-31
35 McKane, M. and Milkman, R. (1995)Genetics. 139, 35-43
| Discussion Board | Previous Page | Your Poster Session |