* Summary
* Introduction
* History
* Report System
* Hardware requirements
*
Operative system
*
Database
*
Local Area Network (LAN)
*
Report System Software (Pathology Information System)
*
Data mining system
*
Activity report (quantification)
* Bibliography
| INABIS '98 Home Page | Your Symposium | Related Symposia & Posters | Scientific Program | Exhibitors' Foyer | Personal Itinerary | New Search |
Management and Automation in Pathology Laboratories
Dr. Marcial García Rojo.
Pathology Dept. Complejo Hospitalario. Ciudad Real. Spain
Information systems in pathology, although frequently integrated in hospital information systems (HIS), are characterized by a very special structure of interface and data, as a consequence of the frequently craft work performed at the laboratory of pathology.
The usual pathway of a specimen –accompanied by its application form with clinical data to be diagnosed in pathology is:
Registry -- Macroscopic study -- Laboratory -- Microscopic study -- Final Diagnosis -- Delivery -- Archives
We will review the impact of new information technologies in each of these steps, analyzing not only the points these technologies can be applied in the generation of pathology reports, but also considering the possibilities in management (i.e. workload), computing assisted diagnosed, communications, and so on.
Before any information system is considered, we find it interesting to review the conclusions of the University of Alabama (http://www.path.uab.edu/), regarding HL7 (Laboratory Medicine Sentinel Monitoring Network: "Test turnaround times, critical value reporting, quality control performance...", completion times, response time, etc.) and the College of American Pathologists Laboratory Accreditation Program http://www.cap.org) that included interest information about configuration of the computing solutions, with referenced to quality control and statistics and considerations about archives (at least 10 years must be recorded).
If we review the history of software applied to pathology in our country, we can find four periods:
1960 – 1980: Big hospitals age. Information systems in mainframes were only found in the most important hospitals (generally more than 800 beds). Basic morphology software in CPM systems was used.
1980 –1990: Informatics departments were created in almost all hospitals. Many of them developed a solution for their pathology department. The commercial solutions were limited, only suitable form medium sized hospitals. New programming techniques appear and a considerable number of pathologists develop their own solutions. Morphology software is becoming more popular and easier to use.
1990-1996: The trend "computing at once" becomes a popular claim between pathologist, since almost a 40% of the pathology departments at that time did not have any information system (not even a personal computer nearby). Some efforts are made in the National Health System to review the information systems available and some basic conclusions are made (Signo project). In these period no new commercial systems appeared, and the need for change in the old systems became apparent in almost all the hospitals. Educational and investigations software appear, with isolated solutions in computing aided diagnosis.
1997-1998: These two years have represented a hallmark in the computing in pathology departments of Spain for two reasons:
a) The active participation of National and Regional Health Systems, that create special committees in order to study the situation. In some Regional Health Systems (Galicia and Catalonia) a information system is designed and developed according to the specifications of their own necessities, and pathology area is part of these systems. In other cases (as in the National Health System) a commercial product is adapted and recommended to use in the hospitals.
b) The greater offer of commercial products. In our review we could find 6 products specifically designed for pathology departments, most of them based in client-server solutions, with integration in HIS, and presenting an easy interface, generally based in 32-bits Windows system.
Apart from report-based systems, the list of software products applied to pathology is increased, not only by some national products (generally multimedia CD-ROMS with educational purposes), but also with all the world-wide solutions nowadays available through Internet.
A recent survey of the Spanish Society of Pathology, reveled:
- A high workload in the secretary staff (3.500 biopsies, 5.300 cytologies and 19 autopsies per year per person)
- 24 % of the pathology information systems is connected to a central HIS.
- Quality control of data is made in a 75% of departments.
Until the end of 1997, the department of pathology of Ciudad Real Hospital, the software used in the report system was based in Clipper for MS-DOS, and developed exclusively for our department. Apart from the technological limitations (and frequent corruption of files when it became larger than 20 MB, for instance), it was an isolated system, disconnected from the HIS, difficult to send a fax from, etc.
As can be concluded in the aforementioned review, nowadays is more complex to make a decision about what software is best for a department, and decisions must be based not only in the design of an attractive multimedia interface, but also in more important items, such as:
- Hardware requirements
- Software maintenance, upgrades, backup, etc.
- Adequacy of data base structure with the already implemented HIS
- Decision between integrated systems and department solutions
- Adaptation to the workflow already established
- Integration of standards in descriptions, diagnoses and codification systems
The renewal of a report system usually is accompanied by a renewal of the hardware equipment. This is a consequence not only of the more exigent requirements of the new software, but also is due to the new specifications in the information systems (local area network connected to HIS, communications by modem/fax, upgrades by FTP, etc.)
Due to the rapid evolution of hardware specifications and the different needs in every department, we don’t think its worth delineating with detail the ideal equipment for each case, but some general rules can be observed:
- Even when we implement the newest technologies, in two years time it can become "old fashioned" in comparison with the new market solutions, but if it was properly sized, in 5 years no major inversion must be needed.
- We must consider two different series of parameters for the motor of our database (the "server") and the client machines. In the server machine, always consider SCSI technology for mass storage (hard disks), in the client side, the new graphics interface is memory consuming, and less than 32 MB of RAM is worthless.
- The local area network must be always Category 5 and fast-ethernet (100 Mbs) must be an objective.
According to the hardware requirements, the server machine will need different operative systems. Multiprocessor machines with high fault-tolerant specifications will require UNIX systems. A medium size hospital (400-700 beds) can find it sufficient to implement a one or (preferable) more Intel-based processor with Windows NT operative systems.
The client-side operative system is always limited to the report software design, and nowadays most solutions are based in Windows 95 and Windows NT platforms.
The most important part of the information system is the database structure and management capabilities (indexation, query system, relational procedures, documental procedures, etc.). Some of these relational database, (Oracle, Informix) were originally designed for UNIX operative systems, but now we can find also a large catalogue of excellent products (Oracle, Informix, Microsoft SQL-Server, Sybase SQL-Anywere, etc.) in the Windows NT platform.
In small departments, relational databases can be based in PC-oriented products (MS-FoxPro, MS-Access, dBase, etc.) bur they are better applied in the client side (interface design).
Independent databases for patients and pathology-related data are important.
The number of personal computers in the pathology department and the kind of information transmitted in the network (text versus image or video predominant) will determine the adequacy of the LAN. Normally a fast-ethernet (100 Mbs) will suffice, but the critical point is the connection with the central HIS that is frequently based in a slow network shared by multiple departments.
Even if our department is based in text-only systems, we think it is interesting to dimension the network to contain also multimedia information, since new technologies (speech recognition, neural networks recognizing patters of images, etc.) will soon require these kind of information to moved from one PC to another in our LAN.
Most database and report software can be used under the TCP/IP standard protocol, and we find it most interesting since these technologies can be then applied to the Internet or Intranet solutions without additional efforts.
Report System Software (Pathology Information System)
The first thing we must consider is the integration of the pathology information system (PIS) in the central hospital information System (HIS). Even when the situation of our department doesn’t allow this integration, we must always consider a change in that situation, which must not require any main change in the department information system. In that way, we find most interesting those systems that allow both an independent existence of the rest of the Hospital (a non desirable situation, but that may be necessary in case of disconnection) and an integration with the HIS.
Normally, the language of programmation used to develop the application is not a relevant decision point in our choice, and it is normally more related to the experience of the software development team than to the excellence of the programming tool. The new object-oriented technologies (C++, PowerBuilder, etc.) are more suitable to develop an client-sever solution that the more higher level languages (Visual Basic, Delphi, dBase, etc.). However, the new models of programmation, including a more distributed environments (DCOM, CORBA, etc.), are introducing new concepts (java applications, java applets, ActiveX controls, XML) that we cannot classify in the older scheme.
Many of the database solutions in the PIS is based in the ODBC standard for data interchange, and many of them will probably be adequate to work in an environment with new emerging interconnectivity standards (Java based connectivity, ActiveX Data Objects or Remote Data Services).
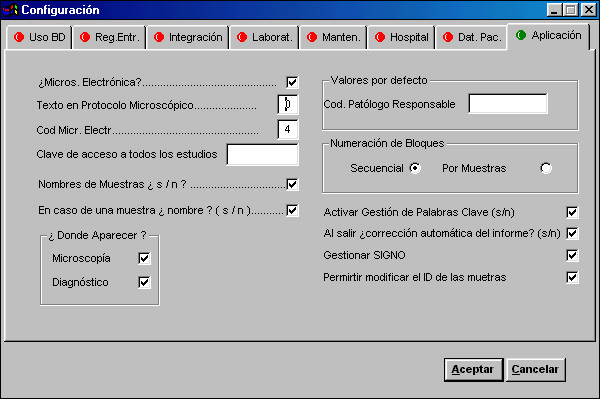
As figure 1 shows, the software must be able to maintain multiple configuration parameters to allow personalization of the system (management of electron microscopy studies, default values, spelling check, quantification study...)
|
The system administrator usually does this only once.
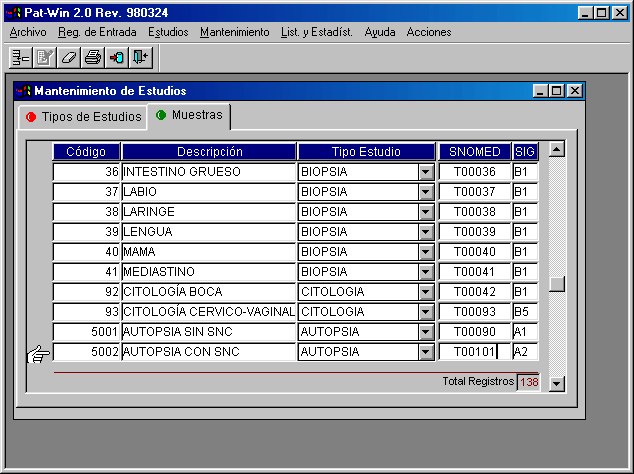
Classification of Specimens
All Studies performed at the pathology department area classified according to a general scheme of Biopsies, Cytology (frequently including fine-needle aspiration biopsies –FNAB-) and Autopsies. Some departments prefer to maintain an independent registry for certain studies (FNAB, immunohistochemistry, electronic microscopy, molecular biology, and son on).
You can see an example at figure 2.
|
Once we make a classification of Type of Studies, we need to define inside each group the types of Specimens we will receive. This will allow us not only to keep descriptions (gross, microscopic, diagnoses) of each specimen. The table containing the different specimens can be adapted to correlate with surgical procedures, but sometimes a list of organs will suffice. Furthermore, the specimen can be associated with a standard classification of topographical or procedural orders (SNOMED) y some value-related classification (we use Signo Project conclusions). These will allow an automated codification and analytical quantification.
We find useful a further classification of specimens relating to the procedure to obtain the specimen (incisional, excisional...: complete resection or only a sample taken in surgical pathology; or exfoliative, imprint, brushing, .... for cytology)
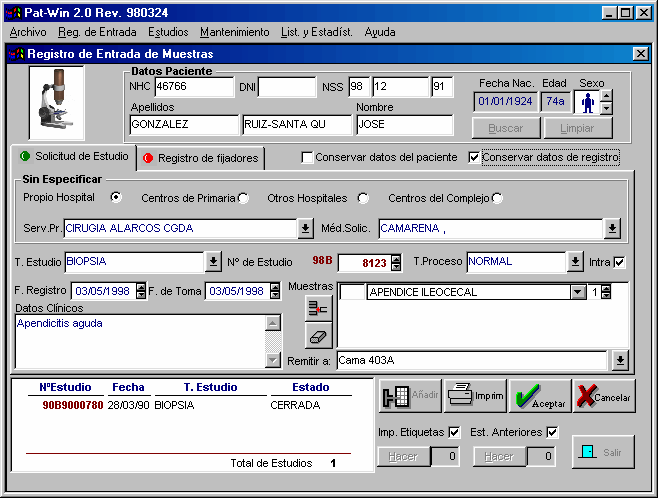
Registry of specimens
It is probable one of the most critical steps, due to the actual difficulties in assigning correctly a specimen to a patient. In some cases the Admission Department is responsible for the database of patients and all the requests are processed here before they are translated to Pathology. But in most cases, it is the secretary of the Pathology Department who has to decide whether a specimen belongs to one patient or not. This is so because most request are incomplete in the affiliation data, and very few requests are received using a code bar system.
This step is therefore oriented to the secretary of the department.
A correlation of data must be established between to ease the entry of data (list of doctors limited to the requesting department, date of registry normally 2 days after date of collections, automatic numbering of specimens and list of previous studies). It is advisable to have an option to maintain all the data from the last patient or the last specimen register, since this will facilitate multiple entries from the same patient of the same requesting department, respectively.
Other options include automatic printing of labels (for bottles of specimens, preferable by code-bar), previous studies or registry book (see figure 3).
|
Intraoperative studies frequently need a special report. At our institution an automated distribution of specimens to pathologist is performed according to the number of registry, but in some institutions the distribution between pathologist depends on topographical data.
In these step it is not always possible to recognize the specimens received (in cytology it may be easier), and we prefer to leave this decision to the next phase.
Gross study
Although partially applicable to cytological and autopsy studies, we will analyze only the gross study of surgical pathology specimens.
Gross study ("carving") allows selecting representative areas of a specimen. Several items are applied in this step:
- Classification: According to topographical, procedural and quantification tables.
- Description: The speech recognition system must be included, but standard pre-formatted texts or protocols must be available. Thus, a simple code ("V1") is all we need to tell the system to complete a standard gall-bladder description.
- Complementary data: Photographs (nowadays simplified with digital cameras), interesting cases record, etc.
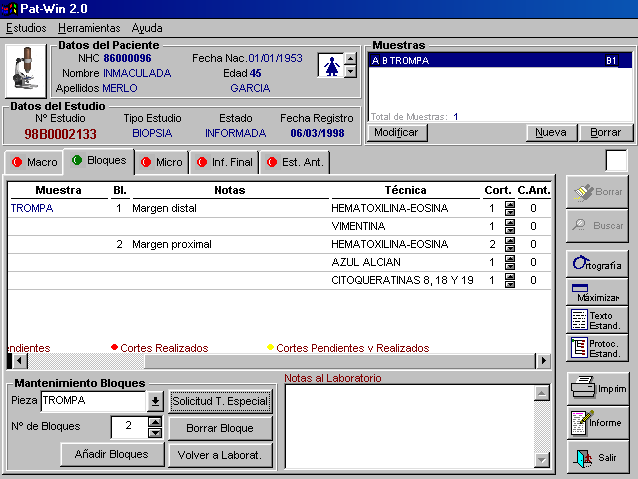
Laboratory
The gross study of a specimen implies the realization of blocks of tissue that must be correctly labeled. In each block one or several stains can be performed. The standard one is hematoxylin-eosin (and the system must know this). Some notes or relevant comments are associated with each block of stain (figure 4).
|
A work list will be created at the laboratory and all completed work will be indicated. Some facilities include printing of labels for slides, grouping of techniques to be performed together, fixatives used. Laboratory techniques can be classified into several laboratories implied (histochemistry, immunochemistry, molecular biology, etc.).
Microscopic study and diagnosis
Once completed the gross study and the staining of all the slides, the pathologist receives these slides, accompanied by the clinical and gross data (printed or accessible at the screen). It is not always possible to access the clinical history from the pathologist’s office, but critical data such as relevant clinical data, gross findings and comments to each block, and list of previous studies, must be accessible on line
Speech recognition and spelling checking are becoming indispensable.
Other possibilities include interest case or tumor committee records, microscopic pictures database, etc.
Coding of diseases must be simplified with the inclusion of topographics once the specimen is grossly studied, and the inclusion of SOMED codes in standard descriptions or standard reports. Although not always easy to implement, it would be interesting the information system to provide an intelligent coding routine based on the final diagnosis.
Standard descriptions and diagnoses
A lot of text must be written in some surgical pathology and anatomic pathology reports. This can be simplified with four tools:
- Speech recognition: After a period of training of 15 days to 1 month (managing vocabulary), low price commercial products (IBM Voice type series) (about 130 – 200 U.S.D.), this systems can be adapted to windows-based PIS.
- Standard descriptions and diagnoses: They can be used directly under the speech recognition system o directly in the pathology information system. We can leave some variables (weight, size, color...) to be filled with proper values.
- Protocols: In staging or grading of tumors it is more advisable to follow a protocol (a series of questions and answers) to obtain a more complete survey. SNOMED codes are automatically included in protocols and standard descriptions.
- Standard reports: Combining protocols and standard descriptions we can make a complete report that will be applied to a study when needed, avoiding all the intermediary steps. In that way, we can tell the editor that this report is an "82E" and a complete cytological report of with hormonal, microbiology, and morphological diagnoses will be generated, and the next to do is just printing it.
The next important aspect in the pathology information system is finding all the data needed with very little effort. Lists and searches must be flexible and may contain administrative data (dates, requesting services, types of studies, pathologists, laboratory staff, age, etc.), pathological data (gross or microscopic findings), diagnoses, coding of diseases (procedures, topography, etiology, etc.).
For that reason, an interesting option is the literal text finding, for example at the text of diagnosis (figure 5), that combined with other fields search (with AND, OR, and NOT Boolean operators) give us unlimited possibilities.
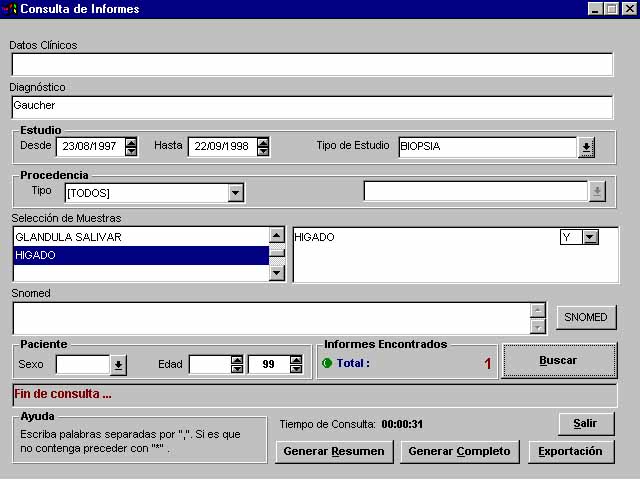 Figure 5. Literal searches combined with administrative and coding matching. |
We must be able to export all the results to standard word processing, spreadsheet or data base software.
Another option must include a list of pictures and photographic slides combined with searches of cases of interest or tumor committees.
A frequent weakness in PIS is the impossibility to perform in only one search a combination of temporal diagnoses, for instance, finding all the patients that first were diagnosed in cytology of "low grade intrasquamous lesion" and afterwards a diagnosis of "high grade intrasquamous lesion" was made.
Activity report (quantification)
The products (specimen) catalogue varies not only from country to country bus also from province to province. We have adopted the nomenclator of the Signo Project, as suggested by the National Health System, although any other classification can be adopted.
Apart from the types of processes, the activity reports also include the number of special techniques (immunohistochemistry, electron microscopy, histochemistry, molecular biology, etc.) performed in each study, as well as pictures taken, and consultation of cases.
We assign a relative value unit (RVU) to each of these parameters.
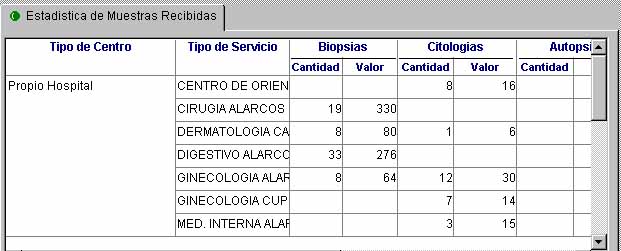
The activity report must include information about requesting departments, types of specimens, including intraoperatory and special techniques, and number of studies and corresponding RVUs (figure 6).
|
Follow up of the studies and turnaround times
The information system usually records the date a study goes to the next phase (for instance, from registry to gross study). Recording of registry, gross, microscopy, laboratory, reported, and closing times are most valuable.
This will allow knowing the exact location of a biopsy or cytological study, and also can be applied to report turnaround times.
We can control the enter and exit times of a request of an ancillary technique, including aspects such as the laboratory staff and kind of cutting to perform.
- Almagro UA, Dunn BE, Choi H, Recla DL. Telepathology. Am J Surg Pathol 1998 Sep;22(9):1161-1163.
- Alvord EC Jr, Siebert JR . Neuropathology: art and science. J Neuropathol Exp Neurol 1997 Dec;56(12):1373
- Cannon TM, Koskela RJ, Burks C, Stallings RL, Ford AA, Hempfner PE, Brown HT, Fickett JW . A program for computer-assisted scoring of Southern blots. Biotechniques 1991 Jun;10(6):764-767
- Cowan DF. Implementing a regulation-complaint quality improvement program on a commercial laboratory information system. Anal Quant Cytol Histol 1992; 14: 407-414.
- Datta A, Rodenacker K . A knowledge acquisition tool in analytical pathology based on multi-media relational database. Comput Methods Programs Biomed 1994 Aug;44(2):119-130
- Della Mea V, Cataldi P, Boi S, Finato N, Della Palma P, Beltrami CA . Image selection in static telepathology through the Internet.J Telemed Telecare 1998;4 Suppl 1:20-22
- Della Mea V, Forti S, Puglisi F, Bellutta P, Finato N, Dalla Palma P, Mauri F, Beltrami CA . Telepathology using Internet multimedia electronic mail: remote consultation on gastrointestinal pathology. .J Telemed Telecare 1996;2(1):28-34
- Erler BS, Chein K, Marchevsky AM. An image analysis workstation for the pathology laboratory Mod Pathol 1993; 6; 612-618.
- Fleege JC, van Diest PJ, Baak JP . Quality control methods for data entry in pathology using a computerized data management system based on an extended data dictionary. Hum Pathol 1992 Feb;23(2):91-97
- Friedman BA. The impact of new features of laboratory information systems on quality assurance in anatomic pathology. Arch Pathol Lab Med 1988 Dec;112(12):1189-1191
- Gil J, Marchevsky AM, Silage DA . Applications of computerized interactive morphometry in pathology: I. Tracings and generation of graphic standards. Lab Invest 1986 Feb;54(2):222-227
- Gonzales J, Cohen MB. Computers in anatomic pathology. A survey of California hospitals. Arch Pathol Lab Med 1988;112:1192-4
- Grasso MA, Ebert D, Finin T . Acceptance of a speech interface for biomedical data collection. Proc AMIA Annu Fall Symp 1997;:739-743
- Gray E, Duvall E, Sprey J, Bird CC . Pathologists dislike sound? Evaluation of a computerised training microscope.J Clin Pathol 1998 Apr;51(4):330-333.
- Horn KD, Sholehvar D, Nine J, Gilbertson J, Hatton C, Richert C, Becich MJ . Continuing medical education on the World Wide Web (WWW). Interactive pathology case studies on the Internet. Arch Pathol Lab Med 1997 Jun;121(6):641-645
- Jelonek J, Stefanowski J. Feature subset selection for classification of histological images. Artif Intell Med 1997 Mar;9(3):227-239.
- Klossa J, Cordier JC, Flandrin G, Got C, Hemet J. A European de facto standard for image folders applied to telepathology and teaching . Int J Med Inf 1998 Feb;48(1-3):207-216
- Komminoth P, Grant JW, Heitz PU. Hochauflosender Video-Graphik-Drucker zur Dokumentation von Operationspraparaten. [A high- resolution video-graphic printer for the documentation of surgical specimens]. Pathologe 1989;10:131-132.
- Larsimont D, Kiss R, d'Olne D, de Launoit Y, Mattheiem W, Paridaens R, Pasteels JL, Verhest A. Relationship between computerized morphonuclear image analysis and histopathologic grading of breast cancer. Anal Quant Cytol Histol 1989 Dec;11(6):433-439
- Lowe HJ, Antipov I, Walker WK, Polonkey SE, Naus GJ. WebReport: a World Wide Web based clinical multimedia reporting system. Proc AMIA Annu Fall Symp 1996: 314-318.
- Lowe HJ, Buchanan BG, Cooper GF, Vries JK . Building a medical multimedia database system to integrate clinical information: an application of high-performance computing and communications technology. Bull Med Libr Assoc 1995 Jan;83(1):57-64.
- McDonald JM, Brossette S, Moser SA. Pathology information systems: data mining leads to knowledge discovery. Arch Pathol Lab Med 1998 May;122(5):409-411
- McDonough E, Cuello BM, Herrera NE. Surgical pathology computerization [letter]. Arch Pathol Lab Med 1985;109:589-590.
- Moffitt ME, Richli WR, Carrasco CH, Wallace S, Zimmerman SO, Ayala AG, Benjamin RS, Chee S, Wood P, Daniels P, et al . MDA-image: an environment of networked desktop computers for teleradiology/pathology. J Med Syst 1991 Apr;15(2):111-115
- Moore GW, Berman JJ. Object-oriented controlled-vocabulary translator using TRANSOFT + HyperPAD. Proc Annu Symp Comput Appl Med Care 1991:973-975.
- Moore GW, Berman JJ. Performance analysis of manual and automated systemized nomenclature of medicine (SNOMED) coding. Am J Clin Pathol 1994; 101: 253-256.
- Moore GW, Berman JJ.Automatic SNOMED coding. Proc Annu Symp Comput Appl Med Care 1994: 225-229.
- Moritz VA, McMaster R, Dillon T, Mayall B . Selection and implementation of a laboratory computer system. Pathology 1995 Jul;27(3):260-267
- Nathwani BN, Heckerman DE, Horvitz EJ, Lincoln TL. Integrated expert systems and videodisc in surgical pathology: an overview. Hum Pathol 1990;21:11-27
- Oberholzer M, Fischer HR, Christen H, Gerber S, Bruhlmann M, Mihatsch M, Famos M, Winkler C, Fehr P, Bachthold L, et al. Telepathology with an integrated services digital network - a new tool for image transfer in surgical pathology: a preliminary report. Hum Pathol 1993; 24: 1078-1085.
- O'Brien MJ, Sotnikov AV, Digital imaging in anatomic pathology. Am J Clin Pathol 1996; 106 (4 Suppl 1): S25-S32
- O'Brien MJ, Takahashi M, Brugal G, Christen H, Gahm T, Goodell RM, Karakitsos P, Knesel EA Jr, Kobler T, Kyrkou KA, Labbe S, Long EL, Mango LJ, McGoogan E, Oberholzer M, Reith A, Winkler C. Digital imagery/telecytology. International Academy of Cytology Task Force summary. Diagnostic Cytology Towards the 21st Century: An International Expert Conference and Tutorial.. Acta Cytol 1998 Jan;42(1):148-164
- Pillarisetti SG. Surgical pathology report in the era of desktop publishing. Arch Pathol Lab Med 1993; 117: 40-42.
- Ranefall P, Wester K, Bengtsson E. Automatic quantification of immunohistochemically stained cell nuclei using unsupervised image analysis. Anal Cell Pathol 1998;16(1):29-43 .
- Rashbass J, Vawer A A networked computer program for managing a national external quality assurance scheme in cytopathology. Cytopathology 1996 Dec;7(6):377-385
- Rothwell DJ, Hause LL. SNOMED and microcomputers in anatomic pathology. Med Inf (Lond) 1983 Jan;8(1):23-31
- Sakurabayashi I [Coding for clinical laboratory information]. [Article in Japanese] Rinsho Byori 1997 Jun;45(6):573-576
- Sasieni PD . The role of statistics in cytopathology. Cytopathology 1997 Jun;8(3):146-147
- Sassi I, Fontanella G, Calandrino R, Faravelli A, Freschi M, Cantaboni A. Descrizione macroscopica di campioni chirurgici assistita da calcolatore. [Computer-assisted macroscopic description of surgical specimens] Pathologica 1986;78:365-369.
- Schubert E, Gross W, Becich MJ. Computer-assisted instruction in pathology residency training: design and implementation of integrated productivity and education workstations. Semin Diagn Pathol 1994 Nov;11(4):282-293
- Simmons MD, Nadin J, Isaac M . Re-engineering pathology: an IT solution to the data conundrum. J Clin Pathol 1997 Oct;50(10):815-818.
- Sociedad Española de Anatomía Patológica. http://www.seap.es/
- Societat Italiana di Anatomia Patologica e Citopatologia Diagnostica. Gestione Anatomia Patologica. http://www.siapec.it/
- Talamo TS, Losos FJ. Surgical pathology accessioning and management on a multiuser hard disk microcomputer system. Arch Pathol Lab Med 1985;109:19-23.
- Taubman SB. A surgical pathology reporting and encoding system using industry- standard hardware and software. Arch Pathol Lab Med 1988;112:1195-1199.
- Teplitz C, Cipriani M, Dicostanzo D, Sarlin J. Automated Speech-recognition Anatomic Pathology (ASAP) reporting. Semin Diagn Pathol 1996; 11: 245-252.
- Tischler AS, Martin MR. Generation of surgical pathology report using a 5,000-word speech recognizer. Am J Clin Pathol 1989;92 (4 Suppl 1):S44-S47.
- True LD . Morphometric applications in anatomic pathology. Hum Pathol 1996 May;27(5):450-467
- Vawer A, Rashbass J . The biological toolbox: a computer program for simulating basic biological and pathological processes. Comput Methods Programs Biomed 1997 Mar;52(3):203-211
- Vuong PN, Vaury P, Balaton AJ, Galet BA, Baviera EE. Numerisation et traitement informatique de l'image: application a l'iconographie des pieces operatoires en anatomie pathologique. [Digitization and image reconstruction: application to iconography of surgical pathology specimens] Ann Pathol; 16: 141-143.
- Wick MR, Archer JB, Isaacs HM, Gross W. Distribution of surgical pathology reports by a computer-driven telephone facsimile (FAX) device. Semin Diagn Pathol 1996; 11: 258-262.
- Wormek AK, Ingenerf J, Orthner HF . SAM: speech-aware applications in medicine to support structured data entry. Proc AMIA Annu Fall Symp 1997;:774-778